Hello! I’d like to ask a noob question: I never used ADQL before, and I am trying to select a random subsample of galaxies from DP0.2. RAND() from standard SQL doesn’t work, and I am having a surprisingly hard time finding how to do this in Google. Gaia tables have a random_idx column, but I haven’t found something like that in DP0.2. Thanks!
Hi @bruno-moraes, great question, thank you for asking!
Not all of the ADQL functionality is yet implemented for queries. I hadn’t tried using RAND() before, so first I confirmed this returns “ERROR Function [RAND] is not found in TapSchema” when I tried to include it in a query. I used the Portal for this test, but it’s the same underlying TAP service in the Portal and Notebook Aspects.
I think the recommended workaround here is to retrieve a larger sample than you need, and then down-select to a random sample. The Object catalog does have an objectId column, but they are not random. Other ideas for generating random samples with DP0.2 are welcome to be posted as replies in this thread.
In the Notebook Aspect, the down-selection could be done with, e.g., options in the numpy.random package (e.g., numpy.random.choice). It is recommended to always include some spatial constraints for queries because the catalogs are so large. Make sure you use a large container when you start your JupyterLab server, when working with large data sets.
In the Portal Aspect, this could be done in a similar way. First do a query, then create a new column using the RAND() function, and then put a limit on the new column to achieve a random subset of the desired size. Below I put a step-by-step simplistic example for doing this.
@bruno-moraes, if this sufficiently answers your question, please mark this post as the solution? And if not, please follow-up in this thread so we can get the issue resolved.
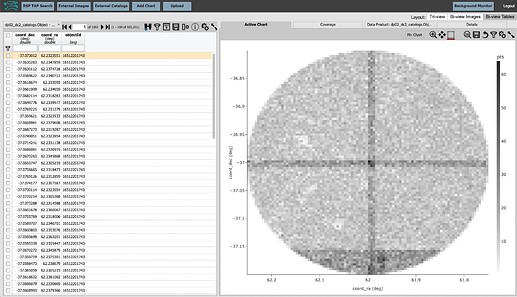
Step-by-step example of creating a random subset of DP0.2 Objects in the Portal Aspect.
Go to data.lsst.cloud and enter the Portal Aspect.
Select “Edit ADQL” from upper right, and copy-paste this query into the ADQL box, or use your own query to obtain only potential galaxies of interest.
SELECT coord_dec,coord_ra,objectId
FROM dp02_dc2_catalogs.Object
WHERE CONTAINS(POINT('ICRS', coord_ra, coord_dec),CIRCLE('ICRS', 62, -37, 0.2))=1
Increase the Row Limit to 200000.
Click Search.
The query returns 105,251 rows.
In the results page, click “Bi-view Tables” at upper right.
The results view screen should now look like this:
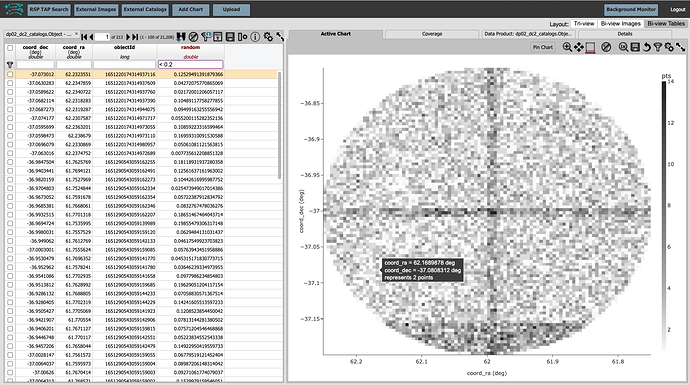
At upper right in the table panel, click on the “Add a column” icon, the vertical rectangle with the plus sign. Hover over icons to see pop-ups of the function.
Name the new column ‘random’ and use the expression ‘RAND()’, like this:
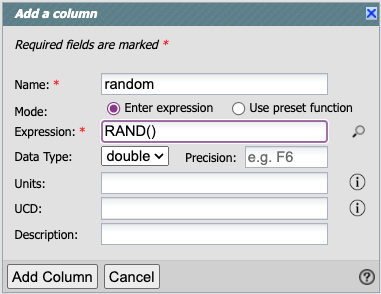
Click “Add Column”.
A new column named random, populated with values between 0 and 1, now appears.
In the box at the top of the column, enter “<0.2” to get a random sample that is 20% the size of the full sample.
This is just a small supplement to Melissa’s answer, regarding her recommendation, “always include some spatial constraints.” If you’d like to use her approach to sample across a big area of the sky, you could stratify by location. Partition the sky into M polygons of equal solid angle. If you want N samples, draw M integers (which will sum to N) from a multinomial distribution in M categories with equal probabilities (or unequal if the solid angles aren’t equal); see scipy.stats.multinomial. Then do the larger-than-needed query in each polygon and downselect to the number from the multinomial. This aims to make sure the samples are appropriately random across the whole area.
Hi @MelissaGraham , @TomLoredo , thanks for the answers! (and apologies for the late reply). I am not 100% these solutions work for us. Our goal is to create a simulated fully-representative spectroscopic training set over the full DP0.2 area and then cut it in colour/magnitude/SNR to imitate certain well-known non-representative spectroscopic samples. We want to run several photo-z trainings and assess the impact of these sample choices. With this in mind:
- using a small area can induce cosmic variance effects;
- the original columns do not allow easily for a SNR cut (or do they?);
I suppose we could stratify by location, but I suppose using a multinomial with fixed total N would oversample low-density regions and undersample high-density ones? Off the top of my head, maybe this would work (adapting Tom’s steps):
- Partition the sky into M polygons.
- Take N_i objects uniformly randomly from each i polygon, where N_i is sampled from a Poisson with fixed_frac * N^tot_i / area_i rate.
Not sure all this is statistically robust, and it would require many many queries to span the footprint, or maybe some smarter cuts on the original sample to reduce its size.
Does this sound overly complicated? Any suggestions?
Thanks!
I wonder if doing 2 separate queries might do the job.
My idea is
- Download all the objectids from the Object table (maybe via an asynchronous query):
query = "SELECT objectId from Object"
results = service.search(query)
- Save the result to a file for general use later:
df = service.search(query).to_table().to_pandas()
df..to_csv('objectIds_all.csv')
- When a subsample is needed, read in the
objectIds_all.csvfile, use numpy to choose a random subsample, and then use the SQL/ADQLWHERE...INfunction to read those random Objects into a table in your current notebook; e.g.:
my_list = "(1249537790362809267, 1252528461990360512, 1248772530269893180, "\
"1251728017525343554, 1251710425339299404, 1250030371572068167, "\
"1253443255664678173, 1251807182362538413, 1252607626827575504, "\
"1249784080967440401, 1253065023664713612, 1325835101237446771)"
query = "SELECT objectId, g_calibFlux, r_calibFlux, i_calibFlux, z_calibFlux "\
"FROM dp02_dc2_catalogs.Object "\
"WHERE objectId IN "+my_list
results = service.search(query)
results.to_table()
(see also ADQL Recipes — Vera C. Rubin Observatory Documentation for Data Preview 0.2).
I’ve found that the SQL/ADQL WHERE...IN function can accept fairly lengthy lists, and, even if not lengthy enough, one could run the commands in Step 3 multiple times with sub-lists until you get the number of random Objects you want.
Hi Douglas,
Thanks for the feedback and suggestion. My student and I will see what we can cook up, sometime in the next few days, and report back.
Cheers,
Bruno
Dear Bruno,
I have some updates that might be useful. These are still in development – and should be treated as such – but I hope they might be helpful!
First, based on suggestions by @MelissaGraham and @leanne, I downloaded all the objectId's from the dp02_dc2_catalogs.Object table. (There are basically a “gazillion” of these ; so this took more effort than I was originally expecting!) I have placed this download in my scratch area on https://data.lsst.cloud; so anyone with an RSP account can access this list without having to re-create it themselves. There is a CSV version and a FITS version:
/scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.csv
/scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.fits
The data in these two files are the same; the only difference is the file format. There are 278,318,455 objectId's in each of these files. They are each about 5GB in size. The FITS file is faster to load than the CSV, but there are some nifty pandas tricks one can use with the CSV file.
I googled around and found this post on “2 Ways to Randomly Sample Rows from a large CSV file”:
https://cmdlinetips.com/2022/07/randomly-sample-rows-from-a-big-csv-file/
I tried the two methods, and they work pretty fast – or at least a lot faster than reading in the whole 5GB CSV file and then using numpy methods to select a random set of objectId's.
First, say you want to grab a random 0.001% of objectId's from the full set listed in /scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.csv. You can use this code snippet to do that:
import pandas as pd
import random
filename = '/scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.csv'
random.seed(4321)
df = pd.read_csv(filename,
skiprows=lambda x: x > 0 and random.random() >=0.00001)
In my case, I got 2872 objectId’s returned.
Or, if you want to grab an exactly N=100 random rows from
/scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.csv, you can use a code snippet like this:
import pandas as pd
import random
filename = '/scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.csv'
sample_n_from_csv(filename, n=100)
where you have previously defined the function sample_n_from_csv as such:
import sys
def sample_n_from_csv(filename:str, n:int=100, total_rows:int=None) -> pd.DataFrame:
if total_rows==None:
with open(filename,"r") as fh:
total_rows = sum(1 for row in fh)
if(n>total_rows):
print("Error: n > total_rows", file=sys.stderr)
skip_rows = random.sample(range(1,total_rows+1), total_rows-n)
return pd.read_csv(filename, skiprows=skip_rows)
One can then feed in that subsample of random objectId's to a WHERE...IN clause of your ADQL query like shown in Step 3 of my previous message, with the caveat that I am not sure what the limit of the number of items in the IN list is (so you might need to break things up into multiple ADQL queries, each with a different sub-batch from the original list of random objectId's.
There may be more updates later, but I hope this helps in case you want to get started before those updates are posted.
Thanks!
Hi @DouglasLTucker, @MelissaGraham ,
Apologies for the belated reply, this is an undergrad project so things move slowly, especially as it is summer vacation here.
TL;DR: Our query adapting Douglas proposed solution works, but it is very slow. It takes 18 minutes to download 2.7k objects. Any bigger query stops with an error after exactly 30m.
Longer explanation: Our solution used the method proposed in the previous answer, selecting a random subsample of the object IDs in the CSV file and using it with a WHERE clause to select the objects from the full table. For the rest of the query, we adapt from Melissa’s example where she created a training set and ran a photo-z algorithm. Our current query looks like this:
SELECT mt.id_truth_type, mt.match_objectId, obj.objectId, obj.detect_isPrimary, ts.ra, ts.dec, ts.redshift, ts.flux_u_noMW, ts.flux_g_noMW, ts.flux_r_noMW, ts.flux_i_noMW, ts.flux_z_noMW, ts.flux_y_noMW
FROM dp02_dc2_catalogs.MatchesTruth AS mt
JOIN dp02_dc2_catalogs.TruthSummary AS ts ON mt.id_truth_type=ts.id_truth_type
JOIN dp02_dc2_catalogs.Object AS obj ON mt.match_objectId=obj.objectId
WHERE obj.objectId IN(1248684569339625874, 1248684569339655215, 1248675773246632823, ...)
AND ts.truth_type=1
AND obj.detect_isPrimary=1
We tested this query with a very small number of objects, and it seems to be working fine. For our purposes, we will refine some of these cuts later. But the issue is that the query is very slow. Running the exact simpler version that Douglas proposed in the previous answer, with no JOINs at all, it takes 18mins to retrieve 2.7k objects. With the JOINS, or trying to run anything bigger, nothing gets past 30 minutes, which makes me think there must be a quota.
We tried using the Large configuration, but we still hit the 30 minutes with a very small number of objects. We don’t know exactly what it means to “use a large container”, as suggested in the first reply to this thread. As for splitting the query in smaller chunks, it is certainly doable, but given current limits, it seems problematic that to get, say, a million objects, we would need to generate hundreds of queries.
So, I guess our next questions are:
- Is it normal that the queries are taking so long to run?
- Is it expected to bump on a 30-min limit? If yes, is it possible to extend it?
- Any suggestions on how to try and improve the performance of the query?
Thanks a lot for your patience and your suggestions!
Best,
Bruno
Hi Bruno – just a note to say the database team has replicated the issue on test systems, and is investigating the performance. Will follow up here when we know more in detail.
–FritzM.
Thanks a lot!
PS. I am taking the liberty of taking the “Solved” tick off for the moment, apologies if this is not good practice.
@bruno-moraes taking the solved tick off IS good practice thank you (I was about to do the same!); opening a new (unsolved) topic would have also been appropriate option.
I’ll have some more advice and a demo as an option for you today.
Hi @bruno-moraes,
Thanks again for following up on this issue. We’ve got a few initial answers and recommendations for you.
First off, the recommendation to “use a large container” means that when presented with the Server Options page during log in to the Notebook Aspect, choose Large in the left-hand menu (probably this is what you meant by large configuration?).
-
Yes, it is my understanding that it is normal at the moment (in the DP0 era) for triple-joins with the truth catalogs to take this long, and that RSP functionality to improve this is still in development.
-
Yes that is expected, as 30 minutes it the current time-out limit imposed on synchronous queries. The solution here is to use an asynchronous query (see below), which can run for much longer. On our side, work to ensure users are provided with an appropriate error message is underway.
-
At the moment, in the DP0 era, I’m not sure there’s much that can be done to speed up a query like this. As I understand it, the data is partitioned by sky coordinate, and this query randomly access many partitions. Future RSP functionality to better support random samples will exist but for now, including some spatial constraints would be the way to go. I didn’t write out an example for this because, based on the above discussion, that sounds like a last resort for you.
If the asynchronous option below works for you please mark this post as the solution instead? As always, follow-ups and feedback are welcome.
Example asynchronous query.
First, the part we’ve already discussed, of importing packages and getting a list of ~2872 random object identifiers.
from lsst.rsp import get_tap_service, retrieve_query
import pandas as pd
import random
service = get_tap_service("tap")
filename = '/scratch/DouglasLTucker/dp02_dc2_catalogs_Object_all_objectids_sorted.csv'
random.seed(4321)
df = pd.read_csv(filename,
skiprows=lambda x: x > 0 and random.random() >=0.00001)
print(len(df))
The use of WHERE <col> IN ()in the query below is a temporary work-around for DP0.2 until the functionality for user-uploaded tables (and joins with user-uploaded tables) is ready.
Prepare the string list of object identifiers. Option to prepare the full list with N = len(df) or first use a short list with N = 100.
N = len(df)
# N = 100
string_list = '('
for i in range(N):
string_list = string_list + str(df['objectId'][i])
if i < N-1:
string_list = string_list + ','
elif i == N-1:
string_list = string_list + ')'
Define the query. Here I’ve used something just really simple as the example, but any query (e.g., your truth table join query) could go here.
query = '''SELECT obj.objectId, obj.detect_isPrimary
FROM dp02_dc2_catalogs.Object AS obj
WHERE obj.objectId IN ''' + string_list
Define the asynchronous query as job. Print the job URL and phase (the URL will be unique to you; the phase will be PENDING).
job = service.submit_job(query)
print('Job URL is', job.url)
print('Job phase is', job.phase)
Start the asynchronous job running.
job.run()
Check on the job phase. It will say “EXECUTING” while the job is in progress, and “COMPLETED” when it is done.
At this time, there is no way to check on the progress or to have a time estimate, or to get a notification when it is done.
print('Job phase is now ', job.phase)
Fetch the results into a table. This will fail with an error if the job phase is not COMPLETED but can be rerun.
async_results = job.fetch_result().to_table().to_pandas()
Hi @bruno-moraes, just wanted to follow up on your ADQL question and to check that @MelissaGraham’s post addressed it. I’m going to go ahead and mark her post as a solution.
FWIW, as we’ve recently got the DP0.2 dataset ingested at the UK-DAC, I was interested to see how our our system preformed here and how to best approach it.
caveat - our qserv system is not as well specced as the qserv system behind the US RSP but we don’t have as many users so …
I used Douglas’s objectId list.
Initially I broke the query up into two queries, one joining Object and MatchesTruth and the other to find the matching records in TruthSummary. I then joined the two astropy results tables i.e the second join was done outside the database.
Using this sampling
skiprows=lambda x: x > 0 and random.random() >=0.01)
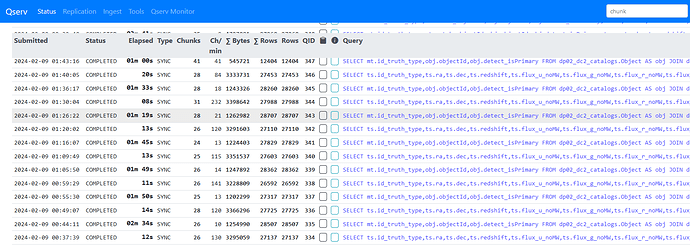
produced just over 2.5 million objectIds. I submitted sync TAP queries with 50,000 objectIds in each initial query. It took around 9hrs to process the 2.5 million objects returning a total of 1.5 million rows matching Bruno’s criteria. Each 50,000 row chunk taking around 10 minutes. I have access to the qserv logs and this shows the queries themselves are only taking around 2 minutes so assume the rest is collating, retrieving and saving the results.
By increasing the total number of objects I think the query efficiency per object is actually improved?
The list is ordered by objectId so increasing the sampling reduces the min/max range of objectIds in a given query and means fewer qserv chunks are accessed. i.e. with the initial sampling producing around 3K objects, these are all submitted in a single query and all 1477 qserv chunks are searched/accessed. With the sampling above, each 50,000 object query was accessing around 25 chunks. Below is a screenshot showing the looped queries, the Object joined with MatchesTruth takes 1-2 minutes, with the follow up TruthSummary query taking 10-20s.
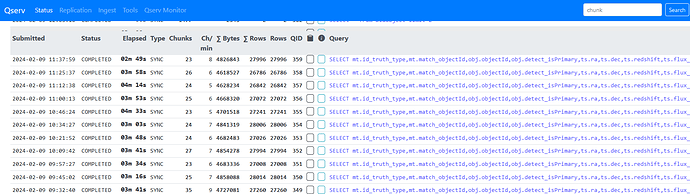
As this approach seemed to work OK, I then tried it as single query with the original 3 table join, again 50,000 objects at a time. These are actually running fine too. A bit slower at around 4 minutes per query, 12 minutes per loop.
I ran the two query approach on the US RSP for 19 chunks and that seems to be doing each 50,000 object chunk in around 14 minutes.
Currently running the single query (three table join) on US RSP and that’s taking about 17 minutes per chunk.
Mike