Hi all,
As explained in this post, I am running ap_pipe on HSC public data to produce light curves, using v20.0.0 and gen2.
I was having issues running it in //, where I had doublets as is also seen here*, as well as other issues like :
InternalError: (psycopg2.errors.InternalError_) right sibling's left-link
doesn't match: block 20306 links to 44760 instead of expected 44023 in index
"PK_DiaSource".
I then decided to run the job in fully sequential mode. This is challenging at CC, since there are no queues to do // computing on a machine with many cores. Instead there are // queues that use MPI, but as far as I understand, ap_pipe can only use python multithreading (with the option -j), but not MPI, as explained here. So in the end I tried to use 16 cores on a single machine on CC, which crashed due to memory limits. I don’t know what can be taking so much memory, and this is something I already mentioned here.
So I am now using 8 cores.
With this configuration, I don’t have any duplicates issue.
I still have RuntimeErrors which completely skip the given ccd, so no sources/objects are added to the db for these.
I had the same failure for the same visits and ccds in my // run. The different errors are:
RuntimeError: Cannot find any objects suitable for KernelCandidacy
RuntimeError: No coadd PhotoCalib found!
RuntimeError: No matches to use for photocal
RuntimeError: Database query failed; did you call make_apdb.py first?
This last error happened to ccds that had worked in my previous // run, so it might be due to some glitch on the server? My job didn’t crash though, and it worked again for the following visits.
Which brings me to my first question:
If I want to run these failed visit/ccd cases again, I will break the “chronology” since I have objects that might have been generated by later visits, to which sources of these earlier visits might be associated. I don’t think this is an issue for my project of generating light curves, but is there any way to fix this, i.e. insert earlier visits that would redefine object ids, re-index etc?
But except for these issues, this is now running ok, without hanging, and without mess in the db. But my main issue is that it is really slow.
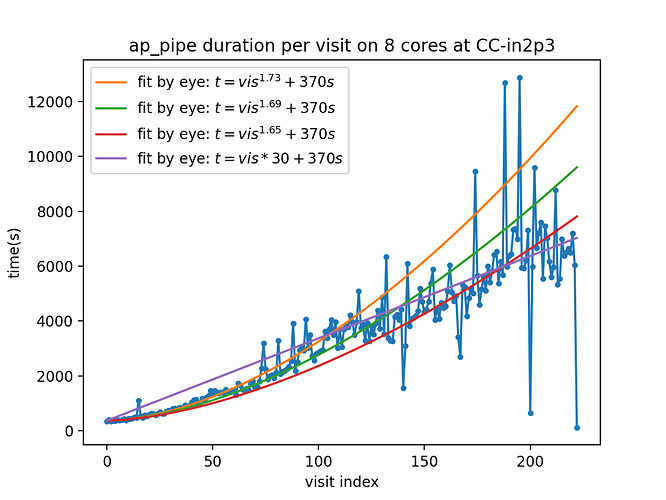
In the figure below, I show the duration of the runs as a function of visit index. I did a few fits by eye in the beginning, which turned out to be quite bad as run went on, but I leave them on the plot.
The few “fast” outliers are jobs that were killed by time limit of my jobs, and restarted, the last one is “in progress”. They currently take about 2h per visit. This is very slow.
Do you know if anyone benchmarked ap_pipe and knows how the time/memory is spent? Association itself should really not take that long, and now, I don’t think there is any weird database access slowdown… Could it be the forced photometry, where I guess it needs to re-access the actual image? I guess it is here updating the forced photometry at every step, whereas I imagine that it only matters at the end, when we have all the sources and a better determined object position…
Is there a way to turn off forced photometry, and only do it in a “post-processing”, or for the last visit?
(Or maybe I miss-understand how forced photometry is done in ap_pipe).
One solution that was suggested by the CC-IN2P3 support, is to use “UNLOGGED TABLES”. I might test this, especially now that I don’t have real clear db issue.
What is the plan in the long term for ap_pipe to deal with the fact that it will get slower and slower as we increase the number of visits? Is it just that for now the code is not fully in place, or that postgresql is not adapted for such datasets?
Sorry for the long post,
Best,
Ben
*though I think my issue was from a different origin: In that DM-26700, the issue was due to a restart of the job after a failure. That happened to me once, but I had duplicates in other situation without job restart, I think due to the fact that I was reading/writting in the database in parallel over the same region of the sky (cosmos).