Hi all,
I am processing DECam data from raw images, utilizing raw object images as well as raw biases and flats. I am trying to produce calibrated CCD images with source catalogs, following pipeline definitions in lsst/obs_decam/pipelines, and using calibration pipelines from lsst/cp_pipe/pipelines.
I am running into an issue running the CharacterizeImageTask to produce sources for the postISRCCD datasets (produced with RunIsrWithCrosstalk.yaml). I have a Gen 3 repo in the directory ./repo, and I am running:
pipetask run \
-j 30 \
-b repo/ \
-i DECam/process/isr,refcats/gen2 \
-o DECam/process/calexp \
-p ProcessCcd.yaml \
--register-dataset-types
where the pipeline definition YAML file is:
# ProcessCcd.yaml
description: ProcessCcd - A set of tasks to run when processing raw images.
instrument: lsst.obs.decam.DarkEnergyCamera
tasks:
characterizeImage:
class: lsst.pipe.tasks.characterizeImage.CharacterizeImageTask
config:
refObjLoader.ref_dataset_name: ps1_pv3_3pi_20170110
The input collection DECam/process/isr is a collection containing postISRCCD datasets:
$ butler query-datasets repo/ --collections DECam/process/isr postISRCCD
type run id band instrument detector physical_filter exposure
---------- ---------------------------------- ---- ---- ---------- -------- -------------------------------- --------
postISRCCD DECam/process/isr/20210430T195740Z 6658 r DECam 1 r DECam SDSS c0002 6415.0 1480.0 989582
postISRCCD DECam/process/isr/20210430T195740Z 6560 r DECam 2 r DECam SDSS c0002 6415.0 1480.0 989582
...
And the input collection refcats/gen2 contains gen2 to gen3 converted Gaia DR1 and Pan-STARRS1 reference catalogs:
$ butler query-datasets repo/ --collections refcats/gen2 gaia_DR1_v1 | head -5
type run id htm8
----------- ------------ ------ -------
gaia_DR1_v1 refcats/gen2 55242 524288
gaia_DR1_v1 refcats/gen2 55243 524289
$ butler query-datasets repo/ --collections refcats/gen2 ps1_pv3_3pi_20170110 | head -10
type run id htm7
-------------------- ------------ ------ ------
ps1_pv3_3pi_20170110 refcats/gen2 530949 131072
ps1_pv3_3pi_20170110 refcats/gen2 530950 131073
ps1_pv3_3pi_20170110 refcats/gen2 530951 131074
ps1_pv3_3pi_20170110 refcats/gen2 530952 131075
ps1_pv3_3pi_20170110 refcats/gen2 530953 131076
ps1_pv3_3pi_20170110 refcats/gen2 530954 131077
ps1_pv3_3pi_20170110 refcats/gen2 530955 131078
Running the pipetask command gives:
$ pipetask run \
> -j 30 \
> -b repo/ \
> -i DECam/process/isr,refcats/gen2 \
> -o DECam/process/calexp \
> -p ProcessCcd.yaml \
> --register-dataset-types
py.warnings WARN: /epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/script/qgraph.py:132: UserWarning: QuantumGraph is empty
qgraph = f.makeGraph(pipelineObj, args)
Traceback (most recent call last):
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/bin/pipetask", line 29, in <module>
sys.exit(main())
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/pipetask.py", line 43, in main
return cli()
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 829, in __call__
return self.main(*args, **kwargs)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 782, in main
rv = self.invoke(ctx)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 1066, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 610, in invoke
return callback(*args, **kwargs)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/decorators.py", line 21, in new_func
return f(get_current_context(), *args, **kwargs)
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/cmd/commands.py", line 102, in run
qgraph = script.qgraph(pipelineObj=pipeline, **kwargs)
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/script/qgraph.py", line 135, in qgraph
raise RuntimeError("QuantumGraph is empty.")
RuntimeError: QuantumGraph is empty.
(lsst-scipipe-0.5.0) bash-4.2$ pipetask run -j 30 -b repo/ -i DECam/process/isr,refcats/gen2 -o DECam/process/calexp -p ProcessCcd.yaml
py.warnings WARN: /epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/script/qgraph.py:132: UserWarning: QuantumGraph is empty
qgraph = f.makeGraph(pipelineObj, args)
Traceback (most recent call last):
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/bin/pipetask", line 29, in <module>
sys.exit(main())
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/pipetask.py", line 43, in main
return cli()
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 829, in __call__
return self.main(*args, **kwargs)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 782, in main
rv = self.invoke(ctx)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 1066, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 610, in invoke
return callback(*args, **kwargs)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/decorators.py", line 21, in new_func
return f(get_current_context(), *args, **kwargs)
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/cmd/commands.py", line 102, in run
qgraph = script.qgraph(pipelineObj=pipeline, **kwargs)
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/script/qgraph.py", line 135, in qgraph
raise RuntimeError("QuantumGraph is empty.")
RuntimeError: QuantumGraph is empty.
(lsst-scipipe-0.5.0) bash-4.2$ pipetask run -j 30 -b repo/ -i DECam/process/isr,refcats/gen2 -o DECam/process/calexp -p ProcessCcd.yaml --register-dataset-types
py.warnings WARN: /epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/script/qgraph.py:132: UserWarning: QuantumGraph is empty
qgraph = f.makeGraph(pipelineObj, args)
Traceback (most recent call last):
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/bin/pipetask", line 29, in <module>
sys.exit(main())
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/pipetask.py", line 43, in main
return cli()
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 829, in __call__
return self.main(*args, **kwargs)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 782, in main
rv = self.invoke(ctx)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 1066, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/core.py", line 610, in invoke
return callback(*args, **kwargs)
File "/astro/users/stevengs/.conda/envs/lsst-scipipe-0.5.0/lib/python3.8/site-packages/click/decorators.py", line 21, in new_func
return f(get_current_context(), *args, **kwargs)
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/cmd/commands.py", line 102, in run
qgraph = script.qgraph(pipelineObj=pipeline, **kwargs)
File "/epyc/projects/decam_ddf/lsst_w_2021_17/stack/miniconda3-py38_4.9.2-0.5.0/Linux64/ctrl_mpexec/21.0.0-26-gb7643ca+642d343ffb/python/lsst/ctrl/mpexec/cli/script/qgraph.py", line 135, in qgraph
raise RuntimeError("QuantumGraph is empty.")
RuntimeError: QuantumGraph is empty.
I think this is telling me that it’s not picking up input data to run on, but I’m not sure.
I’ve tried running CalibrateImageTask within Python on just one exposure that I obtained from the butler myself, and it seems to work:
from lsst.daf import butler as dafButler
from lsst.pipe.tasks.characterizeImage import CharacterizeImageTask, CharacterizeImageConfig
from lsst.pipe.tasks.calibrate import CalibrateTask, CalibrateConfig
import matplotlib.pyplot as plt
import numpy as np
butler = dafButler.Butler("./repo")
registry = butler.registry
postISRCCD_refs = registry.queryDatasets("postISRCCD", collections="DECam/process/isr")
characterize_config = CharacterizeImageConfig()
characterize_config.refObjLoader.ref_dataset_name = "ps1_pv3_3pi_20170110"
characterize_task = CharacterizeImageTask(config=characterize_config)
exposure = butler.get(list(postISRCCD_refs)[0], collections="DECam/process/isr")
characterize_result = characterize_task.run(exposure)
icExp = characterize_result.exposure
icSrc = characterize_result.sourceCat
icSourceSchema = characterize_task.outputSchema.schema
icExpBackground = characterize_result.background
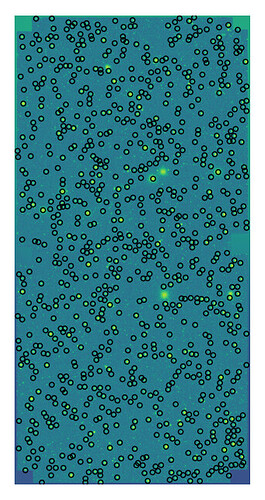
fig = plt.figure(figsize=(8, 4), dpi=200)
plt.imshow(np.arcsinh(icExp.image.getArray()))
plt.scatter(icSrc.getX(), icSrc.getY(), alpha=1, color="k", marker="o", s=5, lw=0.5, facecolors='none')
plt.axis("off")
plt.show()
which produces
Does anyone have a suggestion on how to debug the empty QuantumGraph error when running with pipetask run?