I am working on the effect of LEO Satellites on LSST images. I simulate strong lenses with Lenstronomy, and add mock satellite trails on those. To detect and mask these trails, I want to use maskStreaks module of the LSST Science Pipelines. Since the pipeline functions require full image package (including background, noise, mask layers and instrument information) I import HSC single frame images using the butler and inject the lenses I simulated with streaks. However, I am having trouble with detecting the synthetic streaks using the maskStreaks function. I changed config.nSigma = 0.1 config.invSigma = 0.1 parameters regarding the features of the streaks I simulate but can’t say that it worked. How should I approach to this problem?
Hi @Aysu , could you provide more details about the problem, for example the actual error messages? Also, which version of the LSST Science Pipelines are you using? @mrawls would be a good person to contact for help.
One quick note is that maskStreaks operates on the DETECTED mask plane of the image, so you will have to make sure that is set in your test image. You can do that either by running detection as part of CalibrateImageTask or DetectAndMeasureTask, or by setting the mask plane manually after injecting the synthetic streak.
Hi @sfu, I do not get error messages, I get no results from maskStreaksTask.find(). Recently I revised my streak simulation to get thinner streaks and set nSigma and invSigma to the default values. Now I have a ~60% detection rate. I’m still working on it to get it higher.
@isullivan I figured this out when I checked the source code, so I was doing this
image_array = calexp_image.maskedImage.image.array
streak = ads.create_streak(4176,2048,10e6)
streaky_image = calexp_image.maskedImage.image.array + streak
calexp_image.maskedImage.image.array[:,:] = streaky_image
streaky_mask = calexp_image.maskedImage.mask.array + streak
calexp_image.maskedImage.mask.array[:,:] = streaky_mask
mask_results = mask_streaks_task.find(calexp_image.maskedImage)
mask = mask_results.mask
masked = mask_streaks_task.run(calexp_image.maskedImage)
mask_plane_dict = calexp_image.mask.getMaskPlaneDict()
streak_bit = 1 << mask_plane_dict['STREAK']
mask2 = (calexp_image.maskedImage.mask.array & streak_bit) != 0
masked_image_array1 = calexp_image.maskedImage.image.array.copy()
masked_image_array1[mask2] = np.nan
masked_image_array2 = np.ma.masked_array(calexp_image.maskedImage.image.array,mask)
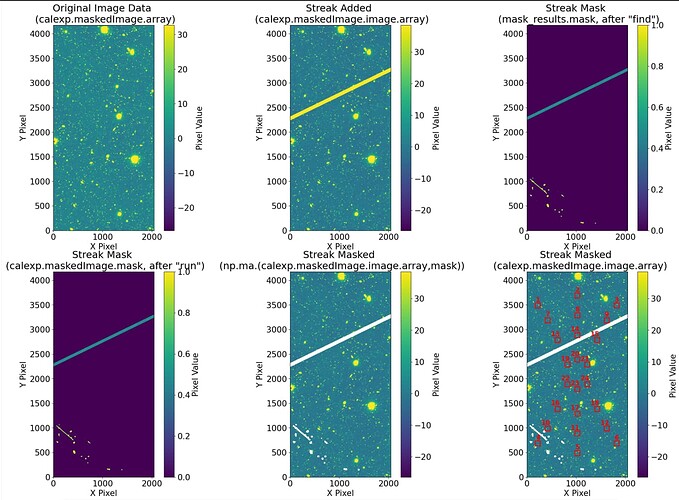
Here, from top right to bottom left:
-
original image from calexp that i call with butler → image_array (calexp_image.maskedImage.image.array)
-
the image after i add the streak → calexp_image.maskedImage.image.array
-
the mask after i run ‘find’, find streak function ->mask (mask_streaks_task.find(calexp_image.maskedImage).mask)
-
the mask after i run ‘run’, mask found streaks function, and extract the streak mask plane and use it to mask
the maskedimage → mask2 (masked = mask_streaks_task.run(calexp_image.maskedImage),
mask_plane_dict = calexp_image.mask.getMaskPlaneDict(),
streak_bit = 1 << mask_plane_dict[‘STREAK’],
mask2 = (calexp_image.maskedImage.mask.array & streak_bit) != 0) -
i manually mask the maskedImage here → masked_image_array2 (np.ma.masked_array(calexp_image.maskedImage.image.array,mask))
-
another way to mask manually by setting streak pixels nan → masked_image_array1( masked_image_array1 = calexp_image.maskedImage.image.array.copy()
masked_image_array1[mask2] = np.nan )
I did not know about the CalibrateImageTask or DetectAndMeasureTask. Should I proceed with them, or is one of the masking methods I used in step/figure 3, 4, 5, and 6 is a valid approach?
Thank you!
If anyone has any recommendations about the correct method, I would appreciate it. Thanks!
Hi @Aysu - I wanted to check to see if you are still having issues with your streak masking. I’ll tag @mrawls again to see if she has any insights that may help.
Hi ! Yes, actually I am still trying to figure our the correct way to detect and mask the synthetic streaks that I inject into HSC images. Thank you!
Hi, thanks for the reminder about this @sgreenstreet.
As Ian pointed out, MaskStreaksTask runs on the DETECTED mask plane. I also note there has been active development on the detection side of streak masking in difference images recently (Nov-Dec 2024).
I suggest you try running detection via DetectAndMeasure[DiaSources]Task (which lives in ip_diffim and calls MaskStreaksTask) with a newer version of the Science Pipelines. Specifically, a new enough ip_diffim to include the changes from DM-45361 and DM-47901 (note the latter also touched meas_algorithms).
If I were to inject streaks, I would first try to use this package, but to be honest your streak looks perfectly streak-y to me, so I suspect focusing your efforts on detection makes the most sense to start.
I understand. Maybe, the reason I got masking with the method I used above is that I was injecting the streak itself into the detected mask plane, so when I used .find or .run it could detect it. I am checking the documentation and the source code for
now. As far as I figured, it needs ‘science, matchedTemplate, difference, scoreExposure’ parameters so I need to get those first I suppose.
Also, I am using calexp from rc2_subset (single frame, g-band) to inject streaks and use these methods, can I consider this dataset a reasonable representative of the HSC observations in general?
Thank you for your answers!
The data you’re using seems reasonable to me. I would suggest doing this as part of a pipeline and not a standalone call to a task so you automatically have all the input connections available. One of the simplest options could be, e.g.,
pipetask run -p $AP_PIPE_DIR/pipelines/HSC/ApPipe.yaml#isr,calibrateImage,retrieveTemplate,subtractImages,detectAndMeasure
(you will also need to specify -d, -i, -o, and -b in the usual fashion). Of note, you will need to provide (an) input collection(s) that include both raws, calibs, and previously built goodSeeingCoadds (for use ase templates) for this pipeline to succeed.
Previously I was simply working on the single frame, after I realized I needed other methods requiring coadds and calibration prior to DetectAndMeasure, as you said, I started to work on the pipeline. Also, I checked the package you recommended in the previous reply, although ‘source injection’ caught my eye in the documentation before I did not realize it could also inject streaks. Maybe I can even try injecting the simulated strong lenses after creating the source injection catalog and pipeline using the same method.
Unrelated, but is there any thread which we can report the parts in the documentation that need to be updated? For example, from Tutorial 3 at gbdes part, when I checked the script #GbdesAstrometricFitTask is now #gbdesAstrometricFit, while cloning rc2_subset branch command for v27 does not exits, etc.
Thank you so much for your time, and suggestions!
Hi @Aysu - As the Forum Watcher this week, I have marked the last response from @mrawls as the solution to the original question regarding streak masking. I have also created a new thread with the question about DP0 documentation updates. If any of this is incorrect, please let me know or please feel free to open a new thread with any additional questions you have.