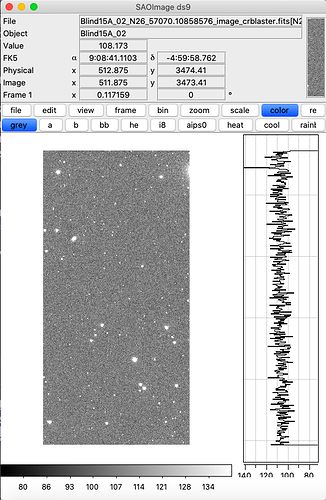
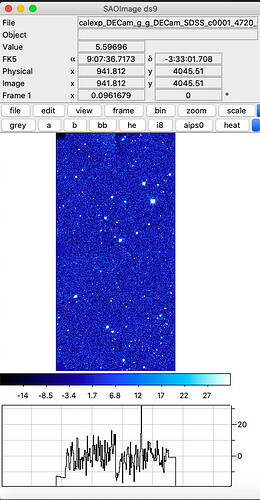
Hi, after doing the step of butler certify-calibration I have been able to make calibrated exposures  , by running the following:
, by running the following:
(lsst-scipipe-0.7.0) [jahumada@leftraru4 jahumada]$ pipetask run -b data_hits/ -p "$AP_PIPE_DIR/pipelines/DarkEnergyCamera/ProcessCcd.yaml" -i DECam/raw/all,refcats,DECam/calib/20140304calibs,hits_master_calib/on20140304_bias,hits_master_calib/on20140304_flats --instrument lsst.obs.decam.DarkEnergyCamera --output-run processCcdOutputs/on20140304_calib_d1 -d "instrument = 'DECam' AND exposure = 289357 AND detector=1" --register-dataset-types -j 2 --pipeline-dot pipeline7.dot
ctrl.mpexec.cmdLineFwk INFO: QuantumGraph contains 3 quanta for 3 tasks, graph ID: '1632843987.2785869-21385'
conda.common.io INFO: overtaking stderr and stdout
conda.common.io INFO: stderr and stdout yielding back
py.warnings WARNING: /mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/obs_base/22.0.1-18-g3ad3863+50f8e256e0/python/lsst/obs/base/formatters/fitsExposure.py:635: UserWarning: Reading file:///mnt/flock/jahumada/data_hits/hits_master_calib/on20140304_flats/20210825T075947Z/flat/g/g_DECam_SDSS_c0001_4720.0_1520.0/flat_DECam_g_g_DECam_SDSS_c0001_4720_0_1520_0_S29_hits_master_calib_on20140304_flats_20210825T075947Z.fits with data ID {instrument: 'DECam', detector: 1, physical_filter: 'g DECam SDSS c0001 4720.0 1520.0', ...}: filter label mismatch (file is None, data ID is FilterLabel(band="g", physical="g DECam SDSS c0001 4720.0 1520.0")). This is probably a bug in the code that produced it.
warnings.warn(f"Reading {self.fileDescriptor.location} with data ID {self.dataId}: "
isr INFO: Converting exposure to floating point values.
isr INFO: Applying crosstalk correction.
isr.crosstalk INFO: Applying crosstalk correction.
isr.crosstalk INFO: Correcting detector S29 with ctSource S28
isr INFO: Assembling CCD from amplifiers.
isr INFO: Applying bias correction.
isr INFO: Applying linearizer.
isr WARNING: Overriding amplifier defined linearityCoeffs ([0 0]) for A
isr WARNING: Overriding amplifier defined linearityCoeffs ([1 0]) for B
isr INFO: Masking defects.
isr INFO: Masking edges as SUSPECT.
isr INFO: Masking non-finite (NAN, inf) value pixels.
isr INFO: Widening saturation trails.
isr INFO: Applying flat correction.
isr INFO: Applying fringe correction after flat.
isr.fringe INFO: Filter not found in FringeTaskConfig.filters. Skipping fringe correction.
isr INFO: Set 457264 BAD pixels to 972.794800.
isr INFO: Interpolating masked pixels.
isr WARNING: No rough magnitude zero point defined for filter g DECam SDSS c0001 4720.0 1520.0.
isr INFO: Setting rough magnitude zero point for filter g DECam SDSS c0001 4720.0 1520.0: 30.940228
isr INFO: Measuring background level.
isr INFO: Flattened sky level: 972.794556 +/- 16.808497.
isr INFO: Measuring sky levels in 8x16 grids: 973.612919.
isr INFO: Sky flatness in 8x16 grids - pp: 0.015974 rms: 0.004328.
ctrl.mpexec.singleQuantumExecutor INFO: Execution of task 'isr' on quantum {instrument: 'DECam', detector: 1, exposure: 289557, ...} took 20.168 seconds
ctrl.mpexec.mpGraphExecutor INFO: Executed 1 quanta successfully, 0 failed and 2 remain out of total 3 quanta.
characterizeImage INFO: PSF estimation initialized with 'simple' PSF
characterizeImage.repair INFO: Identified 40 cosmic rays.
characterizeImage.detection INFO: Detected 71 positive peaks in 50 footprints and 0 negative peaks in 0 footprints to 50 sigma
characterizeImage.detection INFO: Resubtracting the background after object detection
characterizeImage.measurement INFO: Measuring 50 sources (50 parents, 0 children)
characterizeImage.measurePsf INFO: Measuring PSF
characterizeImage.measurePsf INFO: PSF star selector found 26 candidates
characterizeImage.measurePsf.reserve INFO: Reserved 0/26 sources
characterizeImage.measurePsf INFO: Sending 26 candidates to PSF determiner
characterizeImage.measurePsf.psfDeterminer WARNING: NOT scaling kernelSize by stellar quadrupole moment, but using absolute value
characterizeImage.measurePsf INFO: PSF determination using 26/26 stars.
characterizeImage INFO: iter 1; PSF sigma=2.24, dimensions=(41, 41); median background=973.23
characterizeImage INFO: PSF estimation initialized with 'simple' PSF
characterizeImage.repair INFO: Identified 36 cosmic rays.
characterizeImage.detection INFO: Detected 53 positive peaks in 47 footprints and 0 negative peaks in 0 footprints to 50 sigma
characterizeImage.detection INFO: Resubtracting the background after object detection
characterizeImage.measurement INFO: Measuring 47 sources (47 parents, 0 children)
characterizeImage.measurePsf INFO: Measuring PSF
characterizeImage.measurePsf INFO: PSF star selector found 25 candidates
characterizeImage.measurePsf.reserve INFO: Reserved 0/25 sources
characterizeImage.measurePsf INFO: Sending 25 candidates to PSF determiner
characterizeImage.measurePsf.psfDeterminer WARNING: NOT scaling kernelSize by stellar quadrupole moment, but using absolute value
characterizeImage.measurePsf INFO: PSF determination using 25/25 stars.
characterizeImage INFO: iter 2; PSF sigma=2.24, dimensions=(41, 41); median background=973.23
characterizeImage.repair INFO: Identified 37 cosmic rays.
characterizeImage.measurement INFO: Measuring 47 sources (47 parents, 0 children)
characterizeImage.measureApCorr INFO: Measuring aperture corrections for 2 flux fields
characterizeImage.measureApCorr INFO: Aperture correction for base_GaussianFlux: RMS 0.010873 from 24
characterizeImage.measureApCorr INFO: Aperture correction for base_PsfFlux: RMS 0.010428 from 24
characterizeImage.applyApCorr INFO: Applying aperture corrections to 2 instFlux fields
ctrl.mpexec.singleQuantumExecutor INFO: Execution of task 'characterizeImage' on quantum {instrument: 'DECam', detector: 1, visit: 289557, ...} took 18.591 seconds
ctrl.mpexec.mpGraphExecutor INFO: Executed 2 quanta successfully, 0 failed and 1 remain out of total 3 quanta.
calibrate.detection INFO: Detected 207 positive peaks in 201 footprints and 113 negative peaks in 106 footprints to 5 sigma
calibrate.detection INFO: Resubtracting the background after object detection
calibrate.skySources INFO: Added 100 of 100 requested sky sources (100%)
calibrate.deblend INFO: Deblending 301 sources
calibrate.deblend INFO: Deblended: of 301 sources, 6 were deblended, creating 12 children, total 313 sources
calibrate.measurement INFO: Measuring 313 sources (301 parents, 12 children)
calibrate.applyApCorr INFO: Applying aperture corrections to 2 instFlux fields
calibrate INFO: Copying flags from icSourceCat to sourceCat for 39 sources
calibrate.astrometry INFO: Purged 288 sources, leaving 25 good sources
calibrate INFO: Loading reference objects from region bounded by [142.76461127, 143.11502116], [0.72028217, 0.92061049] RA Dec
calibrate INFO: Loaded 2560 reference objects
calibrate WARNING: Reference catalog pm_ra field is not an Angle; cannot apply proper motion.
calibrate WARNING: Found version 0 reference catalog with old style units in schema.
calibrate WARNING: run `meas_algorithms/bin/convert_refcat_to_nJy.py` to convert fluxes to nJy.
calibrate WARNING: See RFC-575 for more details.
calibrate INFO: Converted refcat flux fields to nJy (name, units): (g_flux, ''); (r_flux, ''); (i_flux, ''); (z_flux, ''); (y_flux, ''); (i_fluxSigma, ''); (y_fluxSigma, ''); (r_fluxSigma, ''); (z_fluxSigma, ''); (g_fluxSigma, '')
calibrate.astrometry.referenceSelector INFO: Selected 2560/2560 references
calibrate.astrometry.matcher INFO: Matched 25 sources
calibrate.astrometry.matcher INFO: Matched 25 sources
calibrate.astrometry.matcher INFO: Matched 25 sources
calibrate.astrometry INFO: Matched and fit WCS in 3 iterations; found 25 matches with scatter = 0.021 +- 0.011 arcsec
calibrate.photoCal.match.sourceSelection INFO: Selected 79/313 sources
calibrate INFO: Loading reference objects from region bounded by [142.77040789, 143.09679767], [0.65641708, 0.98277176] RA Dec
calibrate INFO: Loaded 3069 reference objects
calibrate WARNING: Found version 0 reference catalog with old style units in schema.
calibrate WARNING: run `meas_algorithms/bin/convert_refcat_to_nJy.py` to convert fluxes to nJy.
calibrate WARNING: See RFC-575 for more details.
calibrate INFO: Converted refcat flux fields to nJy (name, units): (g_flux, ''); (r_flux, ''); (i_flux, ''); (z_flux, ''); (y_flux, ''); (i_fluxSigma, ''); (y_fluxSigma, ''); (r_fluxSigma, ''); (z_fluxSigma, ''); (g_fluxSigma, '')
calibrate.photoCal.match.referenceSelection INFO: Selected 3069/3069 references
calibrate.photoCal.match INFO: Matched 75 from 79/313 input and 3069/3069 reference sources
calibrate.photoCal.reserve INFO: Reserved 0/75 sources
calibrate.photoCal INFO: Not applying color terms because config.applyColorTerms is None and data is not available and photoRefCat is provided
calibrate.photoCal INFO: Magnitude zero point: 27.878478 +/- 0.000508 from 49 stars
calibrate INFO: Photometric zero-point: 27.878478
calibrate.computeSummaryStats INFO: Measuring exposure statistics
ctrl.mpexec.singleQuantumExecutor INFO: Execution of task 'calibrate' on quantum {instrument: 'DECam', detector: 1, visit: 289557, ...} took 14.933 seconds
ctrl.mpexec.mpGraphExecutor INFO: Executed 3 quanta successfully, 0 failed and 0 remain out of total 3 quanta.
Its quantum graph is:
In the command I ran, I only processed a single ccd. But now when I try the same command for the second detector, I get an error:
(lsst-scipipe-0.7.0) [jahumada@leftraru3 jahumada]$ pipetask run -b data_hits/ -p "$AP_PIPE_DIR/pipelines/DarkEnergyCamera/ProcessCcd.yaml" -i DECam/raw/all,refcats,DECam/calib/20140304calibs,hits_master_calib/on20140304_bias,hits_master_calib/on20140304_flats --instrument lsst.obs.decam.DarkEnergyCamera --output-run processCcdOutputs/on20140304_calib_d2 -d "instrument = 'DECam' AND exposure = 289357 AND detector=2" --register-dataset-types -j 2
ctrl.mpexec.cmdLineFwk INFO: QuantumGraph contains 3 quanta for 3 tasks, graph ID: '1632884828.3815038-54488'
conda.common.io INFO: overtaking stderr and stdout
conda.common.io INFO: stderr and stdout yielding back
py.warnings WARNING: /mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/obs_base/22.0.1-18-g3ad3863+50f8e256e0/python/lsst/obs/base/formatters/fitsExposure.py:635: UserWarning: Reading file:///mnt/flock/jahumada/data_hits/hits_master_calib/on20140304_flats/20210825T075947Z/flat/g/g_DECam_SDSS_c0001_4720.0_1520.0/flat_DECam_g_g_DECam_SDSS_c0001_4720_0_1520_0_S30_hits_master_calib_on20140304_flats_20210825T075947Z.fits with data ID {instrument: 'DECam', detector: 2, physical_filter: 'g DECam SDSS c0001 4720.0 1520.0', ...}: filter label mismatch (file is None, data ID is FilterLabel(band="g", physical="g DECam SDSS c0001 4720.0 1520.0")). This is probably a bug in the code that produced it.
warnings.warn(f"Reading {self.fileDescriptor.location} with data ID {self.dataId}: "
isr INFO: Converting exposure to floating point values.
isr WARNING: Amplifier A is bad.
isr WARNING: Amplifier B is bad.
isr INFO: Applying crosstalk correction.
isr.crosstalk INFO: Applying crosstalk correction.
isr.crosstalk INFO: Correcting detector S30 with ctSource S29
isr INFO: Assembling CCD from amplifiers.
isr INFO: Applying bias correction.
isr INFO: Applying linearizer.
isr WARNING: Overriding amplifier defined linearityCoeffs ([0 0]) for A
isr WARNING: Overriding amplifier defined linearityCoeffs ([1 0]) for B
isr WARNING: 4194304 pixels were out of range of the linearization table
isr WARNING: Amplifier A did not linearize.
isr WARNING: 4194304 pixels were out of range of the linearization table
isr WARNING: Amplifier B did not linearize.
isr INFO: Masking defects.
isr INFO: Masking edges as SUSPECT.
isr INFO: Masking non-finite (NAN, inf) value pixels.
isr WARNING: There were 8388608 unmasked NaNs.
isr INFO: Widening saturation trails.
isr INFO: Applying flat correction.
isr INFO: Applying fringe correction after flat.
isr.fringe INFO: Filter not found in FringeTaskConfig.filters. Skipping fringe correction.
isr INFO: Set 8388608 BAD pixels to nan.
isr INFO: Interpolating masked pixels.
isr WARNING: No rough magnitude zero point defined for filter g DECam SDSS c0001 4720.0 1520.0.
isr INFO: Setting rough magnitude zero point for filter g DECam SDSS c0001 4720.0 1520.0: 30.940228
isr INFO: Measuring background level.
isr INFO: Flattened sky level: nan +/- nan.
py.warnings WARNING: /home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/numpy/core/fromnumeric.py:3419: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
py.warnings WARNING: /home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
py.warnings WARNING: /home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/numpy/core/_methods.py:261: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
py.warnings WARNING: /home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/numpy/core/_methods.py:221: RuntimeWarning: invalid value encountered in true_divide
arrmean = um.true_divide(arrmean, div, out=arrmean, casting='unsafe',
py.warnings WARNING: /home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/numpy/core/_methods.py:253: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
isr INFO: Measuring sky levels in 8x16 grids: nan.
isr INFO: Sky flatness in 8x16 grids - pp: nan rms: nan.
ctrl.mpexec.singleQuantumExecutor INFO: Execution of task 'isr' on quantum {instrument: 'DECam', detector: 2, exposure: 289357, ...} took 13.959 seconds
ctrl.mpexec.mpGraphExecutor INFO: Executed 1 quanta successfully, 0 failed and 2 remain out of total 3 quanta.
ctrl.mpexec.singleQuantumExecutor ERROR: Execution of task 'characterizeImage' on quantum {instrument: 'DECam', detector: 2, visit: 289357, ...} failed
Traceback (most recent call last):
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/singleQuantumExecutor.py", line 174, in execute
self.runQuantum(task, quantum, taskDef, butler)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/singleQuantumExecutor.py", line 450, in runQuantum
task.runQuantum(butlerQC, inputRefs, outputRefs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_tasks/21.0.0-139-g4e3796b1+8c51ef16e5/python/lsst/pipe/tasks/characterizeImage.py", line 342, in runQuantum
outputs = self.run(**inputs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/timer.py", line 181, in wrapper
res = func(self, *args, **keyArgs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_tasks/21.0.0-139-g4e3796b1+8c51ef16e5/python/lsst/pipe/tasks/characterizeImage.py", line 484, in run
background = self.background.run(exposure).background
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/meas_algorithms/22.0.1-21-g532228a4+90bf4c0122/python/lsst/meas/algorithms/subtractBackground.py", line 153, in run
fitBg = self.fitBackground(maskedImage)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/meas_algorithms/22.0.1-21-g532228a4+90bf4c0122/python/lsst/meas/algorithms/subtractBackground.py", line 257, in fitBackground
raise pipeBase.TaskError("All pixels masked. Cannot estimate background")
lsst.pipe.base.task.TaskError: All pixels masked. Cannot estimate background
Process task-1:
Traceback (most recent call last):
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/multiprocessing/process.py", line 315, in _bootstrap
self.run()
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/multiprocessing/process.py", line 108, in run
self._target(*self._args, **self._kwargs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/mpGraphExecutor.py", line 129, in _executeJob
quantumExecutor.execute(taskDef, quantum, butler)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/singleQuantumExecutor.py", line 174, in execute
self.runQuantum(task, quantum, taskDef, butler)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/singleQuantumExecutor.py", line 450, in runQuantum
task.runQuantum(butlerQC, inputRefs, outputRefs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_tasks/21.0.0-139-g4e3796b1+8c51ef16e5/python/lsst/pipe/tasks/characterizeImage.py", line 342, in runQuantum
outputs = self.run(**inputs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/timer.py", line 181, in wrapper
res = func(self, *args, **keyArgs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_tasks/21.0.0-139-g4e3796b1+8c51ef16e5/python/lsst/pipe/tasks/characterizeImage.py", line 484, in run
background = self.background.run(exposure).background
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/meas_algorithms/22.0.1-21-g532228a4+90bf4c0122/python/lsst/meas/algorithms/subtractBackground.py", line 153, in run
fitBg = self.fitBackground(maskedImage)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/meas_algorithms/22.0.1-21-g532228a4+90bf4c0122/python/lsst/meas/algorithms/subtractBackground.py", line 257, in fitBackground
raise pipeBase.TaskError("All pixels masked. Cannot estimate background")
lsst.pipe.base.task.TaskError: All pixels masked. Cannot estimate background
ctrl.mpexec.mpGraphExecutor ERROR: Task <TaskDef(CharacterizeImageTask, label=characterizeImage) dataId={instrument: 'DECam', detector: 2, visit: 289357, ...}> failed; processing will continue for remaining tasks.
ctrl.mpexec.mpGraphExecutor ERROR: Upstream job failed for task <TaskDef(CalibrateTask, label=calibrate) dataId={instrument: 'DECam', detector: 2, visit: 289357, ...}>, skipping this task.
ctrl.mpexec.mpGraphExecutor INFO: Executed 1 quanta successfully, 2 failed and 0 remain out of total 3 quanta.
ctrl.mpexec.mpGraphExecutor ERROR: Failed jobs:
ctrl.mpexec.mpGraphExecutor ERROR: - FAILED: <TaskDef(CharacterizeImageTask, label=characterizeImage) dataId={instrument: 'DECam', detector: 2, visit: 289357, ...}>
ctrl.mpexec.mpGraphExecutor ERROR: - FAILED_DEP: <TaskDef(CalibrateTask, label=calibrate) dataId={instrument: 'DECam', detector: 2, visit: 289357, ...}>
Traceback (most recent call last):
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/bin/pipetask", line 29, in <module>
sys.exit(main())
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cli/pipetask.py", line 52, in main
return cli()
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 829, in __call__
return self.main(*args, **kwargs)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 782, in main
rv = self.invoke(ctx)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 1066, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 610, in invoke
return callback(*args, **kwargs)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/decorators.py", line 21, in new_func
return f(get_current_context(), *args, **kwargs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cli/cmd/commands.py", line 103, in run
script.run(qgraphObj=qgraph, **kwargs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cli/script/run.py", line 171, in run
f.runPipeline(qgraphObj, taskFactory, args)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cmdLineFwk.py", line 669, in runPipeline
executor.execute(graph, butler)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/mpGraphExecutor.py", line 300, in execute
self._executeQuantaMP(graph, butler)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/mpGraphExecutor.py", line 486, in _executeQuantaMP
raise MPGraphExecutorError("One or more tasks failed or timed out during execution.")
lsst.ctrl.mpexec.mpGraphExecutor.MPGraphExecutorError: One or more tasks failed or timed out during execution.
Unfortunately, I’m still unable to retrieve the quantum graph for the latter command because running it again with the --extend-run --skip-existing --pipeline-dot pipeline8.dot, I get:
Traceback (most recent call last):
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 748, in resolveDatasetRefs
raise OutputExistsError(
lsst.pipe.base.graphBuilder.OutputExistsError: Quantum {instrument: 'DECam', detector: 2, visit: 289357, ...} of task with label 'characterizeImage' has some outputs that exist ([DatasetRef(DatasetType('characterizeImage_log', {band, instrument, detector, physical_filter, visit_system, visit}, ButlerLogRecords), {instrument: 'DECam', detector: 2, visit: 289357, ...}, id=a256154a-91ae-4301-a129-fb639c1a33b6, run='processCcdOutputs/on20140304_calib_d2')]) and others that don't ([DatasetRef(DatasetType('icSrc', {band, instrument, detector, physical_filter, visit_system, visit}, SourceCatalog), {instrument: 'DECam', detector: 2, visit: 289357, ...}), DatasetRef(DatasetType('icExp', {band, instrument, detector, physical_filter, visit_system, visit}, ExposureF), {instrument: 'DECam', detector: 2, visit: 289357, ...}), DatasetRef(DatasetType('icExpBackground', {band, instrument, detector, physical_filter, visit_system, visit}, Background), {instrument: 'DECam', detector: 2, visit: 289357, ...}), DatasetRef(DatasetType('characterizeImage_metadata', {band, instrument, detector, physical_filter, visit_system, visit}, PropertySet), {instrument: 'DECam', detector: 2, visit: 289357, ...})]), with no metadata output, and clobbering outputs was not enabled.
Also, I have been able to get some calibrated exposures for other nights, but up until now it only works for the firsts detectors, and no others.
There are also the following cases:
(lsst-scipipe-0.7.0) [jahumada@leftraru3 jahumada]$ pipetask run -b data_hits/ -p "$AP_PIPE_DIR/pipelines/DarkEnergyCamera/ProcessCcd.yaml" -i DECam/raw/all,refcats,DECam/calib/20140302calibs,hits_master_calib/on20140302_bias,hits_master_calib/on20140302_flats --instrument lsst.obs.decam.DarkEnergyCamera --output-run processCcdOutputs/on20140302_calib_d3 -d "instrument = 'DECam' AND exposure = 289149 AND detector=3" --register-dataset-types
Traceback (most recent call last):
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/bin/pipetask", line 29, in <module>
sys.exit(main())
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cli/pipetask.py", line 52, in main
return cli()
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 829, in __call__
return self.main(*args, **kwargs)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 782, in main
rv = self.invoke(ctx)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 1066, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/core.py", line 610, in invoke
return callback(*args, **kwargs)
File "/home/jahumada/.conda/envs/lsst-scipipe-0.7.0/lib/python3.8/site-packages/click/decorators.py", line 21, in new_func
return f(get_current_context(), *args, **kwargs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cli/cmd/commands.py", line 102, in run
qgraph = script.qgraph(pipelineObj=pipeline, **kwargs)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cli/script/qgraph.py", line 148, in qgraph
qgraph = f.makeGraph(pipelineObj, args)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/ctrl_mpexec/22.0.1-18-gee15f3f+b3a3f94acb/python/lsst/ctrl/mpexec/cmdLineFwk.py", line 569, in makeGraph
qgraph = graphBuilder.makeGraph(pipeline, collections, run, args.data_query, metadata=metadata)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 938, in makeGraph
return scaffolding.makeQuantumGraph(metadata=metadata)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 835, in makeQuantumGraph
graph = QuantumGraph({task.taskDef: task.makeQuantumSet() for task in self.tasks}, metadata=metadata)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 835, in <dictcomp>
graph = QuantumGraph({task.taskDef: task.makeQuantumSet() for task in self.tasks}, metadata=metadata)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 361, in makeQuantumSet
return set(q.makeQuantum() for q in self.quanta.values())
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 361, in <genexpr>
return set(q.makeQuantum() for q in self.quanta.values())
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/graphBuilder.py", line 262, in makeQuantum
helper.adjust_in_place(self.task.taskDef.connections, self.task.taskDef.label, self.dataId)
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/connections.py", line 699, in adjust_in_place
adjusted_inputs_by_connection, adjusted_outputs_by_connection = connections.adjustQuantum(
File "/mnt/flock/jahumada/lsstw_v21_37/lsst-w.2021.37/scripts/stack/miniconda3-py38_4.9.2-0.7.0/Linux64/pipe_base/22.0.1-23-g91f4350+66b3ef8762/python/lsst/pipe/base/connections.py", line 582, in adjustQuantum
raise FileNotFoundError(
FileNotFoundError: Not enough datasets (0) found for non-optional connection calibrate.astromRefCat (ps1_pv3_3pi_20170110) with minimum=1 for quantum data ID {instrument: 'DECam', detector: 3, visit: 289149, ...}.
I suppose this error could be straightforward to solve by downloading and ingesting the corresponding Pan-STARRS images.
If you have any idea of what could be happening, please let me know 
Thanks for the help!


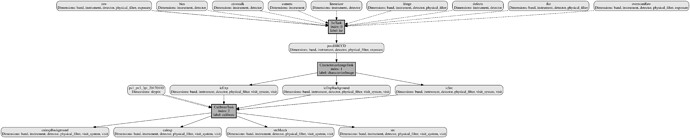
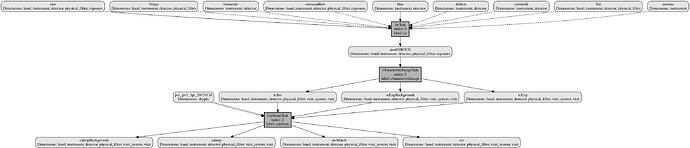
 . Maybe I should start again with a new REPO? I’m not sure how to clear the collections and products I already have with the ‘old’ configuration.
. Maybe I should start again with a new REPO? I’m not sure how to clear the collections and products I already have with the ‘old’ configuration.