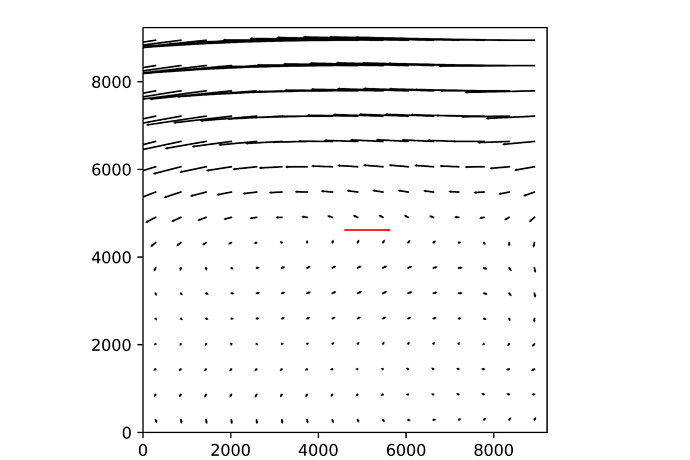
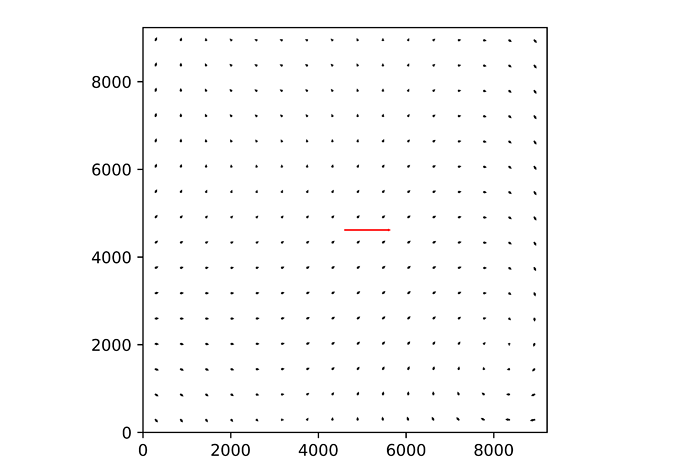
Recently I run your pipeline to calibrate a TAN-SIP distortion of the mock data, and it works pretty good:
for example it’s match radius converges to 0.01 pixel after only 1 iter:
<match radius> = 40.4 +- 14.87 [74 matches]
Dropping into debugger to allow inspection of display. Type 'continue' when done.
(Pdb) continue
<match radius> = 0.01142 +- 0.07599 [74 matches]
Dropping into debugger to allow inspection of display. Type 'continue' when done.
And the resiudals of the image is:
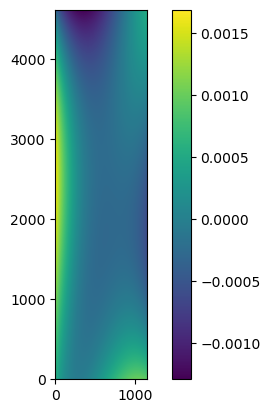
And I want to know how many stars are necessary for one image(see what will happen if the match number become less), In the above output log, it’s match number is [74 matches]
And I try to achieve the aim by set:
config.astrometry.matcher.numBrightStars = 20
which will infect the max_n_patters in pessimistic_pattern_matcher_b_3D.py(After read the code I think it’s the config I should to modify but it didn’t work.)
So I try to trim the source catalog by modify the code
goodSourceCat = sourceCat
# to
goodSourceCat = sourceCat[:30]
in matchPessimisticB.py , it actually reduce the number of match, but here the sourcecatalog is not sorted, it will affect the steps after that.
so I modify the
sorted_source_array = source_array[source_array[:, -1].argsort(), :3]
# to
sorted_source_array = source_array[source_array[:, -1].argsort(), :3][:30]
in pessimistic_pattern_matcher_b_3D.py, but it didn’t work.
So, where should I modify?
Another question:
in the config file of HSC
# Better astrometry matching
config.astrometry.matcher.numBrightStars = 150
with a comment # Better astrometry matching, why the value 150 is better than others? how it’s been confirmed?
Thank you!