That is weird. As far as I can tell, all the overrides are applied before anything is done to create the database, so order should not matter.
I see, then I’m not sure what could’ve gone wrong in my make_apdb.py command before…  I’m just glad that it works now
I’m just glad that it works now 
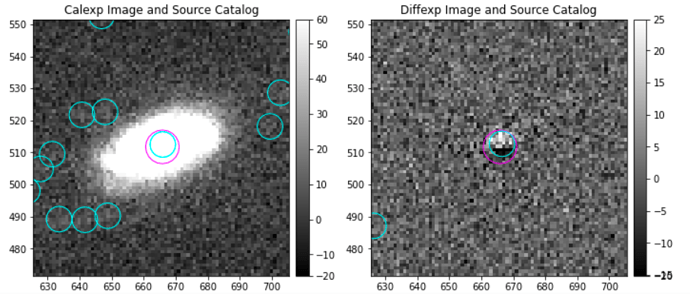
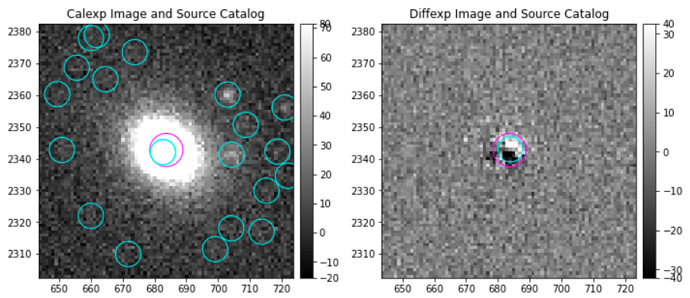
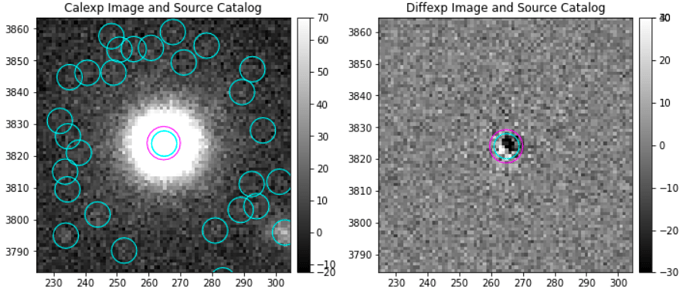
Hello! I have a question about the image differencing task. I have been able to obtain image-differenced images, and now I’m measuring the fluxes of my sources of interest. What I’ve noticed is that an important percentage of them show ‘bipolar’ residuals, which apparently are caused by a somewhat shifted subtraction between the calibrated exposure and the template (deep Coadd). Here are some examples of what I’m talking about:
I wonder if there is a method to better this subtraction, such as a geometric correction of some sort?
PS: here is an example of the command line I’ve been using to make image-differencing images:
pipetask run -b data_hits -i Coadd_AGN/Blind15A_16,processCcdOutputs/Blind15A_16,refcatsv2,DECam/raw/all,DECam/calib,skymaps,DECam/calib/20180503calibs,DECam/calib/20150219calibs,DECam/calib/20150220calibs,DECam/calib/20150218calibs,DECam/calib/20150217calibs,DECam/calib/20150221calibs -p $AP_PIPE_DIR/pipelines/DarkEnergyCamera/ApPipe.yaml --output-run imagDiff_AGN/Blind15A_16 -d "exposure IN (411011, 410961, 410905, 411446, 411396, 411245, 411295, 411345, 412050, 412297, 412240,742493, 742490, 742494, 742489) AND detector in (35, 49, 16, 24, 30, 41, 46)" -j 3 -c diaPipe:apdb.db_url='sqlite:////home/jahumada/databases/blind15a_16.db' -c diaPipe:apdb.isolation_level='READ_UNCOMMITTED' --register-dataset-types
Hi! Recently I tried implementing ZOGY with the LSST Science Pipeline (ZogyImagePsfMatchTask — LSST Science Pipelines) and have also encountered some issues with the resulting images. Here’s an example:

The magenta circle has a radii of 1 arcsec
As you see with this source, there seems to be some error within the pipeline. The sharp and rectangular edges suggest that maybe the geometrical correction/warping of the template image is not ideal. Maybe there is a configuration for DECam images in particular that could solve this? For now, I’m just using the run() function and putting as input the calibrated exposure as science and its corresponding coadd as the template exposure.
Happy to read any comments/suggestions ![]()
Hi Paula, I don’t think anyone on the Science Pipelines team has worked much with the ZOGY algorithm recently. I realize I linked to it a while back, but you might find the summary page from last year’s image differencing sprint interesting: Image Differencing Audit. It might help to compare the template and science images side-by-side, as well as looking at what the difference image looks like using default configs vs. using ZOGY.
Hi Meredith, thanks!
I think I need permission or an account to see the summary you linked. How can I have access to this?
Ah, I forgot all our confluence pages require a login, my bad! Here is a PDF-ification of the summary in question.
sullivan-ImageDifferencingAudit-180522-2115-340.pdf (79.6 KB)
Thanks a lot, Meredith! that report helps a lot as a guide ![]()
I’m trying some of the configurations and checking which one looks best for my images.
I have a question regarding one of the plots from the jupyter notebook. These types of plots specifically:
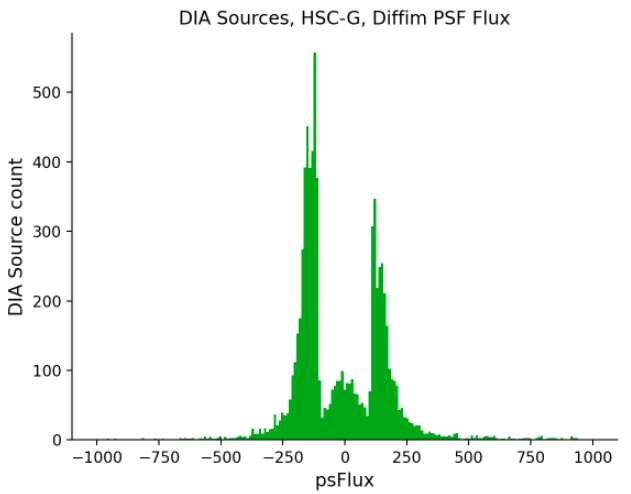
Is each bin of the histogram the flux measured of a source with a PSF aperture? or could it be the sum of different flux profiles from the DIA sources?.
Thanks a lot !
Paula.
Several kinds of fluxes are measured…
- psFlux from DIA Source table (plotted here): forced PSF photometry on diffim at DIA Source position
- totFlux from DIA Source table: forced PSF photometry on calexp at DIA Source position
- apflux from DIA Source table: aperture photometry on diffim at DIA Sources position
- psFlux from DIA Forced Source Table: forced PSF photometry on diffim at DIA Object position
- totFlux from DIA Forced Source Table: forced PSF photometry on calexp at DIA Object position
There are also, of course, fluxes in the src catalog (produced alongside calexps), which result from PSF photometry on the calexp at the calexp-determined source position.
Thanks for the description Meredith ![]()
Right now I’m trying image differencing with the configuration of pre-convolution on the science image, and for some of the ccds the run seems to fail…
It appears in the output that there is some kind of bug that produces this:
py.warnings WARNING: /home/jahumada/lsstw_v22_10/lsst-w.2022.10/stack/miniconda3-py38_4.9.2-2.0.0/Linux64/obs_base/geff6ec3125+d22c7905a5/python/lsst/obs/base/formatters/fitsExposure.py:651: UserWarning: Reading file:///home/jahumada/data_hits/hits_master_calib/20150221_flats/20211221T232825Z/flat/g/g_DECam_SDSS_c0001_4720.0_1520.0/flat_DECam_g_g_DECam_SDSS_c0001_4720_0_1520_0_N21_hits_master_calib_20150221_flats_20211221T232825Z.fits with data ID {instrument: 'DECam', detector: 52, physical_filter: 'g DECam SDSS c0001 4720.0 1520.0', ...}: filter label mismatch (file is None, data ID is FilterLabel(band="g", physical="g DECam SDSS c0001 4720.0 1520.0")). This is probably a bug in the code that produced it.
warnings.warn(
The warning indicates that there’s a filter label mismatch… I’m not sure how this can be solved.
Thanks!
It looks like you encountered this in a previous community post ![]()
I know @lskelvin previously had this issue when processing some DECam data last year, maybe he remembers the specific resolution?
This ticket is related, but as far as I can tell, it’s aimed at fixing an annoying warning, and doesn’t seem to be stopping image processing from happening.
Are you sure there’s not some other error in your traceback unrelated to this warning message?
Hi Paula,
I can confirm that the recently merged ticket that @mrawls linked to above does address the filter label mismatch warning issue that you discovered. The calibration combination task was not previously copying filter information from raw files into newly constructed calibration files. This issue has now been resolved by @czw and should no longer impact future data reductions. With that said however, any calibration files which have previously been constructed (e.g., flats, sky frames) will need to be regenerated in order to stop this warning from appearing.
The good news is that this warning should not impact your reductions at all (beyond causing annoyance). The warning you noticed is an indication that the filter information is being fixed at runtime.
I haven’t had a chance to catch up on all of the discussion in this thread, but I hope this helps somewhat? Keen to hear how you get on!
Thanks for all your comments ![]() I will eventually update the software, and hope to avoid this warning!.
I will eventually update the software, and hope to avoid this warning!.
Right now I have a greater issue, and it regards running the AP Pipe on different configurations. I’ve attempted running image differencing with the following configurations:
doDecorrelation : TruedoPreConvolve : True- default
What worries me is that the resulting images look the same for all configurations! I’m not sure why… here are some stamps of a source of interest:
The first row are the calibrated images of the same source at different epochs, and the second row are the differenced-images.
J095753.59+015831.3_doDec2_stamps.pdf (220.4 KB)
J095753.59+015831.3_PreConv_stamps.pdf (220.5 KB)
J095753.59+015831.3_stamps.pdf (216.4 KB)
These configurations I add them directly in the ApPipe.yaml file, under the ImageDifference config section. Maybe I’m missing something here?
Thanks!
I took a quick look at this. Default A&L differencing sets doDecorrelation to True, so that explains why your first case matches your third case. In addition, it appears that as of about a year ago, doPreConvolve was deprecated and now one must set useScoreImageDetection instead - this explains your second case.
If it’s any consolation, we are very close to completing a long-overdue full refactor of imageDifferenceTask! You can see the details on DM-33001. That ticket is “Done,” but we have not yet adopted the new task(s) in our default AP or DRP pipelines - testing and validation is still in progress.
I see!
Great. I will try this out ![]()
Thanks a lot!
Hi! I encountered an error that I’m not sure how to work around.
setting useScoreImageDetection as True will rise the following error:
ValueError: doWriteSubtractedExp=True and useScoreImageDetection=True Regular difference image is not calculated. AL subtraction calculates either the regular difference image or the score image.
but setting useScoreImageDetection as True and doWriteSubtractedExp as False results in an empty QuantumGraph:
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'Coadd_AGN/Blind15A_16_2exp'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'processCcdOutputs/Blind15A_16'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'refcatsv2'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/raw/all'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'skymaps'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib/20180504calibs'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib/20150219calibs'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib/20150220calibs'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib/20150218calibs'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib/20150217calibs'.
lsst.pipe.base.graphBuilder WARNING: No datasets of type goodSeeingDiff_differenceExp in collection 'DECam/calib/20150221calibs'.
py.warnings WARNING: /home/jahumada/lsstw_v22_10/lsst-w.2022.10/stack/miniconda3-py38_4.9.2-2.0.0/Linux64/ctrl_mpexec/g34d403d65c+f0cf790be1/python/lsst/ctrl/mpexec/cli/script/qgraph.py:183: UserWarning: QuantumGraph is empty
qgraph = f.makeGraph(pipelineObj, args)
lsst.daf.butler.cli.utils ERROR: Caught an exception, details are in traceback:
Traceback (most recent call last):
File "/home/jahumada/lsstw_v22_10/lsst-w.2022.10/stack/miniconda3-py38_4.9.2-2.0.0/Linux64/ctrl_mpexec/g34d403d65c+f0cf790be1/python/lsst/ctrl/mpexec/cli/cmd/commands.py", line 126, in run
qgraph = script.qgraph(pipelineObj=pipeline, **kwargs)
File "/home/jahumada/lsstw_v22_10/lsst-w.2022.10/stack/miniconda3-py38_4.9.2-2.0.0/Linux64/ctrl_mpexec/g34d403d65c+f0cf790be1/python/lsst/ctrl/mpexec/cli/script/qgraph.py", line 186, in qgraph
raise RuntimeError("QuantumGraph is empty.")
RuntimeError: QuantumGraph is empty.
Not sure how I should proceed ![]() , I bet there’s a parameter that I should consider, but I don’t know which.
, I bet there’s a parameter that I should consider, but I don’t know which.
Thanks!
I find it a bit weird that both of these parameters cannot be True at the same time. I understand that doWriteSubtractedExp writes/registers the difference images…
I haven’t played much with these specific configs and don’t have any insights, sorry. I think the newly refactored ImageDifference I mentioned before is probably going to be easier to switch to than sorting out the idiosyncrasies of every possible config combination in “old” ImageDifference. Is there something in particular that isn’t working how you’re hoping for with the default configs?
I’m currently comparing the light curves I get from Image Differencing versus the light curves obtained by aperture photometry for the same sources. I expect both of these to have a similar structure, that is, the same tendencies.
Unfortunately, there are many cases where both of these LCs have a significantly different nature, and it so happens to be for the sources where the subtraction is worse. With the latter, I mean that there are big flux dipoles.
I was hoping that doing pre-convolution to the science images may help with these dipoles.
I assume that I can access this refactored ImageDifference with the newest release of the pipeline? ![]()
In principle yes, though we are still testing it ![]() you can follow along in semi-real-time here. Very heavy disclaimers, there are almost certainly some bugs we haven’t squashed yet - in particular, the science image pre-convolution option is giving us some weird patch edge artifacts at this moment. I defer to @kherner for specific pointers he may wish to give. It may be best to give us a bit to iron out the rough parts of science image pre-convolution before diving in.
you can follow along in semi-real-time here. Very heavy disclaimers, there are almost certainly some bugs we haven’t squashed yet - in particular, the science image pre-convolution option is giving us some weird patch edge artifacts at this moment. I defer to @kherner for specific pointers he may wish to give. It may be best to give us a bit to iron out the rough parts of science image pre-convolution before diving in.