Hi everyone!
In DP0, I used the AlardLuptonSubtractTask to do image subtraction for a custom pipeline. I based it on the example in DP0 tutorial 14. However, it seems my code is not working anymore in DP1, and I can’t seem to figure out why. I also couldn’t find a DP1 tutorial on the RSP that covers image subtraction, so I was hoping I could find an answer here. Below is my old code (that worked in DP0 in 2024_42):
# Load data
calexp = butler.get('calexp', dataId=dataId)
template = butler.get('goodSeeingDiff_templateExp', dataId=dataId)
sources = butler.get('src', dataId=dataId)
# Do the subtraction
config = AlardLuptonSubtractConfig()
config.sourceSelector.value.unresolved.name = 'base_ClassificationExtendedness_value'
alTask = AlardLuptonSubtractTask(config=config)
diff = alTask.run(template, science, sources)
To run with DP1, I changed the first few lines to
calexp = butler.get('visit_image', dataId=dataId)
template = butler.get('template_coadd', dataId=dataId)
sources= butler.get('source', dataId=dataId)
But when I then do alTask.run(template, science, sources), I get the following error:
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
Cell In[22], line 4
2 config.sourceSelector.value.unresolved.name = 'base_ClassificationExtendedness_value'
3 alTask = AlardLuptonSubtractTask(config=config)
----> 4 diff = alTask.run(template, calexp, sources_calexp)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/utils/g8fa50ef3f3+fcb1d3bbc8/python/lsst/utils/timer.py:308, in timeMethod.<locals>.decorator_timer.<locals>.timeMethod_wrapper(self, *args, **keyArgs)
299 logInfo(
300 obj=self,
301 prefix=func.__name__ + "Start",
(...)
305 stacklevel=stacklevel,
306 )
307 try:
--> 308 res = func(self, *args, **keyArgs)
309 finally:
310 logInfo(
311 obj=self,
312 prefix=func.__name__ + "End",
(...)
316 stacklevel=stacklevel,
317 )
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/ip_diffim/g8b8da53e10+8f7b08dc1c/python/lsst/ip/diffim/subtractImages.py:373, in AlardLuptonSubtractTask.run(self, template, science, sources, visitSummary)
331 @timeMethod
332 def run(self, template, science, sources, visitSummary=None):
333 """PSF match, subtract, and decorrelate two images.
334
335 Parameters
(...)
371 set, is less then the configured requiredTemplateFraction
372 """
--> 373 self._prepareInputs(template, science, visitSummary=visitSummary)
375 # In the event that getPsfFwhm fails, evaluate the PSF on a grid.
376 fwhmExposureBuffer = self.config.makeKernel.fwhmExposureBuffer
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/ip_diffim/g8b8da53e10+8f7b08dc1c/python/lsst/ip/diffim/subtractImages.py:954, in AlardLuptonSubtractTask._prepareInputs(self, template, science, visitSummary)
938 def _prepareInputs(self, template, science, visitSummary=None):
939 """Perform preparatory calculations common to all Alard&Lupton Tasks.
940
941 Parameters
(...)
952 lookup.
953 """
--> 954 self._validateExposures(template, science)
955 if visitSummary is not None:
956 self._applyExternalCalibrations(science, visitSummary=visitSummary)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/ip_diffim/g8b8da53e10+8f7b08dc1c/python/lsst/ip/diffim/subtractImages.py:788, in AlardLuptonSubtractTask._validateExposures(template, science)
769 @staticmethod
770 def _validateExposures(template, science):
771 """Check that the WCS of the two Exposures match, the template bbox
772 contains the science bbox, and that the bands match.
773
(...)
786 bounding box, or if the bands do not match.
787 """
--> 788 assert template.wcs == science.wcs, \
789 "Template and science exposure WCS are not identical."
790 templateBBox = template.getBBox()
791 scienceBBox = science.getBBox()
AssertionError: Template and science exposure WCS are not identical.
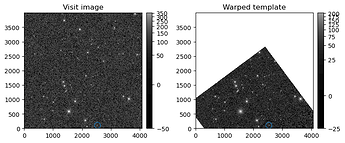
This suggests that the template is no longer aligned with the calexp. After plotting both the calexp and template, this is indeed the case. I don’t remember that this was the case in DP0.2. Is there no longer a corresponding template for each calexp/visit_image? Are we supposed to align the templates ourselves now, or did I somehow load the template incorrectly?
I ignored this error for now by just duplicating the science exposure and injecting a source in the duplicate, and hoping to subtract those two. But I quickly ran into another error:
-----------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[26], line 4
2 config.sourceSelector.value.unresolved.name = 'base_ClassificationExtendedness_value'
3 alTask = AlardLuptonSubtractTask(config=config)
----> 4 diff = alTask.run(inj_calexp, calexp, sources_calexp)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/utils/g8fa50ef3f3+fcb1d3bbc8/python/lsst/utils/timer.py:308, in timeMethod.<locals>.decorator_timer.<locals>.timeMethod_wrapper(self, *args, **keyArgs)
299 logInfo(
300 obj=self,
301 prefix=func.__name__ + "Start",
(...)
305 stacklevel=stacklevel,
306 )
307 try:
--> 308 res = func(self, *args, **keyArgs)
309 finally:
310 logInfo(
311 obj=self,
312 prefix=func.__name__ + "End",
(...)
316 stacklevel=stacklevel,
317 )
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/ip_diffim/g8b8da53e10+8f7b08dc1c/python/lsst/ip/diffim/subtractImages.py:449, in AlardLuptonSubtractTask.run(self, template, science, sources, visitSummary)
447 sourceMask = science.mask.clone()
448 sourceMask.array |= template[science.getBBox()].mask.array
--> 449 selectSources = self._sourceSelector(sources, sourceMask)
450 if convolveTemplate:
451 self.metadata["convolvedExposure"] = "Template"
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/ip_diffim/g8b8da53e10+8f7b08dc1c/python/lsst/ip/diffim/subtractImages.py:868, in AlardLuptonSubtractTask._sourceSelector(self, sources, mask)
844 def _sourceSelector(self, sources, mask):
845 """Select sources from a catalog that meet the selection criteria.
846
847 Parameters
(...)
865 remaining after source selection.
866 """
--> 868 selected = self.sourceSelector.selectSources(sources).selected
869 nInitialSelected = np.count_nonzero(selected)
870 selected *= self._checkMask(mask, sources, self.config.excludeMaskPlanes)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/meas_algorithms/g72a202582f+7a25662ef1/python/lsst/meas/algorithms/sourceSelector.py:720, in ScienceSourceSelectorTask.selectSources(self, sourceCat, matches, exposure)
718 selected &= self.config.flags.apply(sourceCat)
719 if self.config.doUnresolved:
--> 720 selected &= self.config.unresolved.apply(sourceCat)
721 if self.config.doSignalToNoise:
722 selected &= self.config.signalToNoise.apply(sourceCat)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/share/eups/Linux64/meas_algorithms/g72a202582f+7a25662ef1/python/lsst/meas/algorithms/sourceSelector.py:453, in RequireUnresolved.apply(self, catalog)
437 def apply(self, catalog):
438 """Apply the flag requirements to a catalog
439
440 Returns whether the source is selected.
(...)
451 (True means selected).
452 """
--> 453 value = catalog[self.name]
454 return BaseLimit.apply(self, value)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/lib/python3.12/site-packages/astropy/table/table.py:2093, in Table.__getitem__(self, item)
2091 def __getitem__(self, item):
2092 if isinstance(item, str):
-> 2093 return self.columns[item]
2094 elif isinstance(item, (int, np.integer)):
2095 return self.Row(self, item)
File /opt/lsst/software/stack/conda/envs/lsst-scipipe-10.0.0/lib/python3.12/site-packages/astropy/table/table.py:264, in TableColumns.__getitem__(self, item)
253 """Get items from a TableColumns object.
254
255 ::
(...)
261 tc[1:3] # <TableColumns names=('b', 'c')>
262 """
263 if isinstance(item, str):
--> 264 return OrderedDict.__getitem__(self, item)
265 elif isinstance(item, (int, np.integer)):
266 return list(self.values())[item]
KeyError: 'base_ClassificationExtendedness_value'
I believe this error comes from the fact that sources is now a table, whereas before it was a SourceCatalog. Perhaps butler.get('source', dataId=dataId) is not the DP1 equivalent of butler.get('src', dataId=dataId)?
I would love to hear if anyone else has managed to get image subtraction to work. Any feedback or ideas to solve either of the two issues above is much appreciated ![]() Thanks!
Thanks!
Cheers,
Tobias