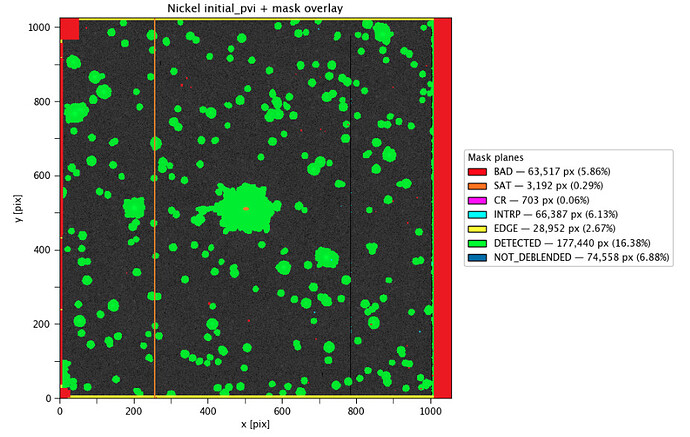
I’m in the process of debugging my obs package for the Nickel telescope and custom processCcd pipeline for the Nickel Telescope, and I’ve been running into a persistent issue where the overscan region isn’t trimmed in downstream datasets.
Currently I have my obs package running with a basic single frame pipeline consisting of ISR + CalibrateImage steps. I’m working on some finer tuning aspects (defect mask, color terms, etc), but it’s successfully detecting sources and running astrometry / photometry.
It is my understanding that post ISR datasets should have the overscan region fully trimmed out of the image. Yet despite specifying the overscan region in my camera yaml, I’m still seeing the overscan region in postISRCCD and initial_pvi dataset exposures (in addition to flat, bias, etc). I’m also The overscan region appears to be masked as BAD pixels, but it’s definitely still there.
Is this expected behavior? Or am I dealing with overscan incorrectly?
For context, the Nickel CCD has an overscan region in the rightmost 32 columns. I’m also masking ~30 data columns at the rightmost edge of the frame (to the left of the overscan region) to deal with what I’ve determined to be rolloff.
I’m including some visualizations and obs_nickel code snippets below that may be helpful.
camera/nickel.yaml
name: Nickel
# --- Amplifier Template ---
AMP: &
perAmpData: true
dataExtent: [1056, 1024] # full raw frame
readCorner: LL
rawBBox: [[0, 0], [1056, 1024]]
rawDataBBox: [[0, 0], [1025, 1024]] # Everything left of overscan is signal
rawSerialPrescanBBox: [[0, 0], [0, 0]] # No prescan
rawSerialOverscanBBox: [[1025, 0], [31, 1024]] # Right-side overscan
rawParallelPrescanBBox: [[0, 0], [0, 0]] # No paralell prescan
rawParallelOverscanBBox: [[0, 0], [0, 0]] # No parallel overscan
gain: 1.8
readNoise: 10.7
saturation: 65535
linearityType: PROPORTIONAL
linearityThreshold: 0
linearityMax: 65535
linearityCoeffs: [0.0, 1.0]
# --- CCD Template ---
CCD: &CCD
detectorType: 0 # SCIENCE
refpos: [528.0, 512.0] # center of 1056 x 1024
offset: [0.0, 0.0]
bbox: [[0, 0], [1056, 1024]]
pixelSize: [0.015, 0.015] # 15 micron pixels
transformDict:
nativeSys: 'Pixels'
transforms: {}
transposeDetector: false
pitch: 0.0
yaw: 0.0
roll: 0.0
amplifiers:
A00:
<<: *AMP
hdu: 0
ixy: [0, 0]
flipXY: [false, false]
# --- Global Focal Plane Scale ---
plateScale: 24.7 # arcsec/mm = 0.37 arcsec/pixel ÷ 0.015 mm
# --- Optical Distortion Approximation ---
transforms:
nativeSys: 'FocalPlane'
FieldAngle:
transformType: radial
coeffs: [0.0, 1.0, 0.0]
# --- Detector Table ---
CCDs:
CCD0:
<<: *CCD
id: 0
name: CCD0
serial: Nickel-1
physicalType: SCIENCE
refpos: [528.0, 512.0]
offset: [0.0, 0.0, 0.0]
amplifiers:
A00:
<<: *AMP
gain: 1.8
readNoise: 10.7
hdu: 0
ixy: [0, 0]
flipXY: [false, false]
Pipeline Shell Script
#!/usr/bin/env bash
# bad exposures - exclude:
BAD="1032,1033,1043,1046,1047,1048,1049,1050,1051,1052,1056"
########## ABSOLUTE PATHS (edit if needed) ##########
REPO="/Users/dangause/Desktop/lick/lsst/data/nickel/062424"
RAWDIR="/Users/dangause/Desktop/lick/data/062424/raw"
OBS_NICKEL="/Users/dangause/Desktop/lick/lsst/lsst_stack/stack/obs_nickel"
REFCAT_REPO="/Users/dangause/Desktop/lick/lsst/lsst_stack/stack/refcats"
########## BASIC CONFIG ##########
INSTRUMENT="lsst.obs.nickel.Nickel"
RUN="Nickel/raw/all"
TS="$(date -u +%Y%m%dT%H%M%SZ)"
echo "=== Nickel pipeline starting @ $TS ==="
########## CREATE & REGISTER ##########
if [ ! -f "$REPO/butler.yaml" ]; then
butler create "$REPO"
fi
butler register-instrument "$REPO" "$INSTRUMENT" || true
butler ingest-raws "$REPO" "$RAWDIR" --transfer symlink --output-run "$RUN"
butler define-visits "$REPO" Nickel
########## CURATED ##########
CURATED="Nickel/run/curated/$TS"
butler write-curated-calibrations "$REPO" Nickel "$RUN" --collection "$CURATED"
########## BIAS ##########
CP_RUN_BIAS="Nickel/run/cp_bias/$TS"
pipetask run \
-b "$REPO" \
-i "$CURATED","$RUN" \
-o "$CP_RUN_BIAS" \
-p "$CP_PIPE_DIR/pipelines/_ingredients/cpBias.yaml" \
-d "instrument='Nickel' AND exposure.observation_type='bias'" \
--register-dataset-types
# certify bias broadly
butler certify-calibrations "$REPO" "$CP_RUN_BIAS" "$CURATED" bias \
--begin-date 2020-01-01 --end-date 2030-01-01
########## FLATS ##########
CP_RUN_FLAT="Nickel/run/cp_flat/$TS"
pipetask run \
-b "$REPO" \
-i "$CURATED","$RUN","$CP_RUN_BIAS" \
-o "$CP_RUN_FLAT" \
-p "$CP_PIPE_DIR/pipelines/_ingredients/cpFlat.yaml" \
-c cpFlatIsr:doDark=False \
-d "instrument='Nickel' AND exposure.observation_type='flat'" \
--register-dataset-types
########## DEFECTS (from flats; module has no detector args) ##########
DEF_TS="$(date -u +%Y%m%dT%H%M%SZ)"
DEFECTS_RUN="Nickel/calib/defects/$DEF_TS"
QA_DIR="$OBS_NICKEL/scripts/defects/qa_$DEF_TS"
python "$OBS_NICKEL"/scripts/defects/make_defects_from_flats.py \
--repo "$REPO" \
--collection "$CP_RUN_FLAT" \
--register \
--ingest \
--defects-run "$DEFECTS_RUN" \
--plot \
--qa-dir "$QA_DIR"
# === AUTO-PICK LATEST DEFECTS RUN ===
DEFECTS_RUN="$(butler query-collections "$REPO" | awk '/^Nickel\/calib\/defects\//{print $1}' | tail -n1)"
echo "Using latest defects run: $DEFECTS_RUN"
# point current -> latest defects
butler collection-chain "$REPO" Nickel/calib/defects/current "$DEFECTS_RUN" --mode redefine
########## UNIFIED CALIB CHAIN ##########
CALIB_CHAIN="Nickel/calib/current"
butler collection-chain "$REPO" "$CALIB_CHAIN" \
"$CURATED" "$CP_RUN_BIAS" "$CP_RUN_FLAT" Nickel/calib/defects/current \
--mode redefine
########## REFCATS (run from refcat repo; original commands) ##########
cd "$REFCAT_REPO"
# Gaia DR3
convertReferenceCatalog \
data/gaia-refcat/ \
scripts/gaia_dr3_config.py \
./data/gaia_dr3_all_cones/gaia_dr3_all_cones.csv \
&> convert-gaia.log
butler register-dataset-type "$REPO" gaia_dr3_20250728 SimpleCatalog htm7
butler ingest-files \
-t direct \
"$REPO" \
gaia_dr3_20250728 \
refcats/gaia_dr3_20250728 \
data/gaia-refcat/filename_to_htm.ecsv
butler collection-chain \
"$REPO" \
--mode extend \
refcats \
refcats/gaia_dr3_20250728
# PS1 DR2
convertReferenceCatalog \
data/ps1-refcat/ \
scripts/ps1_config.py \
./data/ps1_all_cones/merged_ps1_cones.csv \
&> convert-ps1.log
butler register-dataset-type "$REPO" panstarrs1_dr2_20250730 SimpleCatalog htm7
butler ingest-files \
-t direct \
"$REPO" \
panstarrs1_dr2_20250730 \
refcats/panstarrs1_dr2_20250730 \
data/ps1-refcat/filename_to_htm.ecsv
butler collection-chain \
"$REPO" \
--mode extend \
refcats \
refcats/panstarrs1_dr2_20250730
########## SCIENCE PROCESSING ##########
cd "$OBS_NICKEL"
PIPE="$OBS_NICKEL/pipelines/ProcessCcd.yaml"
PROCESS_CCD_RUN="Nickel/run/processCcd/$(date +%Y%m%dT%H%M%S)"
# quick sanity
butler query-collections "$REPO" | grep -E 'Nickel/calib/(current|defects/current)' || true
pipetask run \
-b "$REPO" \
-i "$RUN","$CALIB_CHAIN","refcats" \
-o "$PROCESS_CCD_RUN" \
-p "$PIPE#processCcd" \
-d "instrument='Nickel' AND exposure.observation_type='science' AND NOT (exposure IN (${BAD}))" \
--register-dataset-types \
2>&1 | tee logs/processCcd_$TS.log
# -d "instrument='Nickel' AND exposure.observation_type='science'" \
echo "=== Done ==="
echo "Curated: $CURATED"
echo "CP Bias: $CP_RUN_BIAS"
echo "CP Flat: $CP_RUN_FLAT"
echo "Defects run: $DEFECTS_RUN"
echo "Calib chain: $CALIB_CHAIN"
echo "Science run: $PROCESS_CCD_RUN"
processCcd.yaml
# pipelines/ProcessCcd.yaml
description: ISR + Image Calibration
tasks:
isr:
class: lsst.ip.isr.IsrTask
config:
doOverscan: True
doBias: True
doFlat: True
doDark: False
doDefect: True
doFringe: False
doSuspect: True
doWrite: True
doTrimToMatchCalib: True
doVignette: True
fluxMag0T1:
B: 2e8
V: 2e8
R: 2e8
I: 2e8
calibrateImage:
class: lsst.pipe.tasks.calibrateImage.CalibrateImageTask
config:
connections.astrometry_ref_cat: gaia_dr3_20250728
connections.photometry_ref_cat: panstarrs1_dr2_20250730
astrometry.forceKnownWcs: False
# match search‐radius: 60″ ≃ 300 pix @ 0.2″/pix
astrometry.matcher.maxOffsetPix: 300
astrometry.matcher.maxRotationDeg: 1.0
astrometry.matcher.matcherIterations: 12
# accept a sloppy initial fit up to 60″ mean, then refine
astrometry.maxMeanDistanceArcsec: 60.0
astrometry.matchDistanceSigma: 8.0
astrometry.maxIter: 12
# astrometry_ref_loader.anyFilterMapsToThis: None
# astrometry_ref_loader.filterMap: {"b": "gMeanPSFMag", "v": "gMeanPSFMag", "r": "rMeanPSFMag", "i": "iMeanPSFMag", "u": "gMeanPSFMag"}
photometry_ref_loader.filterMap: {"b": "gMeanPSFMag", "v": "gMeanPSFMag", "r": "rMeanPSFMag", "i": "iMeanPSFMag", "u": "gMeanPSFMag"}
# photometry_ref_loader.filterMap: {"b": "g", "v": "g", "r": "r", "i": "i", "u": "g"}
photometry.match.matchRadius: 60.0
subsets:
processCcd:
- isr
- calibrateImage
steps:
- label: processCcd
sharding_dimensions: visit,detector