Thank you very much for these resources. They have been very useful for understanding more about Butler.
Nevertheless, I still have some problems ingesting non-LSST images into a Butler repository. I wasn’t able to create a metadata translator for the instrument (I was using some VST OmegaCAM and FORS2 images) but I will keep studying to understand how it works (I have little experience with Python so I need a bit more time).
Beyond that, I think I found another way of running pipeline tasks on FITS images even though it is probably far from being the best solution. Despite this, it seems to work at least for what I had in mind, so I share it here, probably it could be helpful for someone else too.
Details here
Essentially, what I have done is using the readFits method of the ExposureF class:
import lsst.afw.image as afwImage
image = afwImage.exposure.ExposureF.readFits('myImage1.fits')
where 'myImage1.fits' is the relative path of my FITS image. It complains about the mask which I think is not fatal:
lsst.afw.image.MaskedImageFitsReader WARNING: Expected extension type not found: IMAGE
but for the rest, it seems to work. I was able to do a cut of the image:
import lsst.geom as afwGeom
x_target, y_target = 1000, 500
width,height=600,500
xmin,ymin = x_target-width//2, y_target-height//2
bbox = afwGeom.Box2I()
bbox.include(afwGeom.Point2I(xmin, ymin))
bbox.include(afwGeom.Point2I(xmin+width, ymin+height))
cutout = image.Factory(image, bbox, origin=afwImage.LOCAL, deep=False)
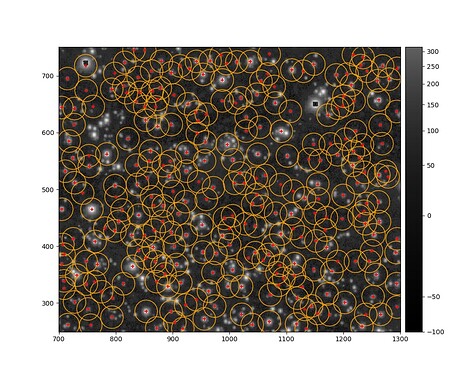
Additionally, I was able to run some pipeline tasks such as:
from lsst.pipe.tasks.characterizeImage import CharacterizeImageTask
config = CharacterizeImageTask.ConfigClass()
config.psfIterations = 1
charImageTask = CharacterizeImageTask(config=config)
result = charImageTask.run(cutout)
and finally, I get the sources identified and overplotted on the original image: