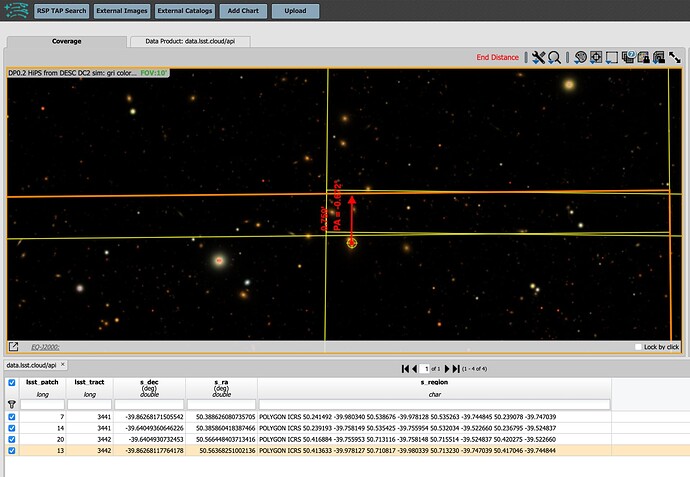
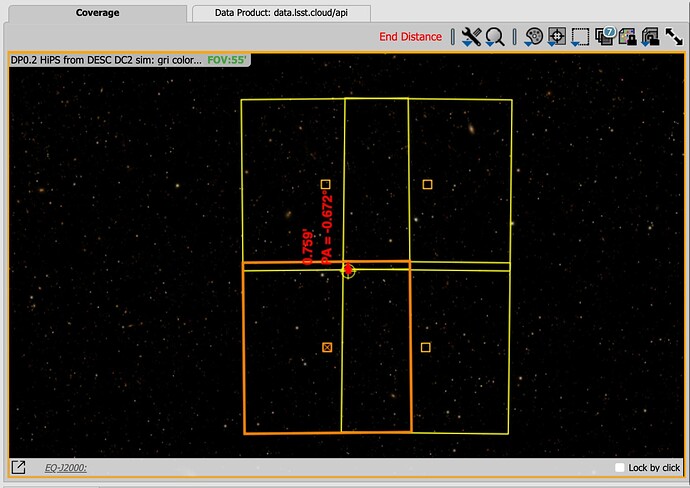
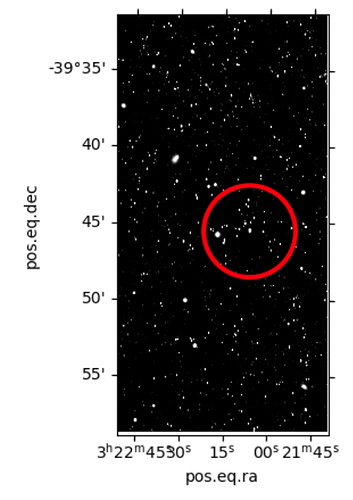
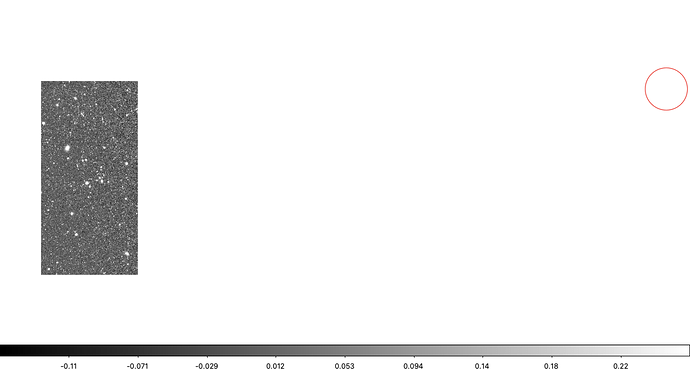
Hello again, I have a (bit of a hack) method for using GetTemplateTask with patches from two different tracts, instead of the same tract. It comes down to defining the bounding box. That’s the part that wasn’t working.
I found that bigImageBBox = nImage1.getBBox().expandedTo(nImage2.getBBox()) didn’t produce an appropriate bounding box. Image 2 is east of Image 1, but the bigImageBBox gets expanded to positive X pixels. When GetTemplateTask is used to project Image 2 into bigImageBBox, there are no pixels at the location of the Image 2 data. At least, that’s my interpretation of what’s happening.
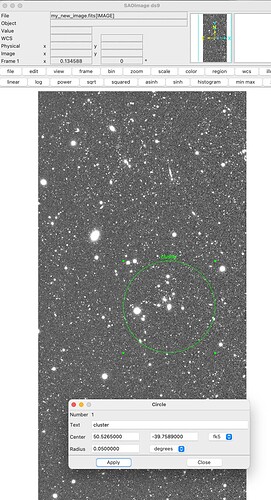
Below, a bigImageBBox is defined by hand to be what I estimated as an approximately appropriate expansion of Image 1 to allow for pixels from Image 2. In the image below the code snippet, it seems I might have underestimate the lower extent of bigImageBBox and cut off a bit of the bottom left, but, at least this gives the idea. And the patch/tract boundary looks pretty seamless.
import lsst.afw.display as afwDisplay
from lsst.daf.butler import Butler
from lsst.ip.diffim import GetTemplateTask
from lsst.afw.image import Image
from lsst.geom import Box2I, Point2I, Extent2I
import matplotlib.pyplot as plt
import numpy as np
afwDisplay.setDefaultBackend('matplotlib')
config = 'dp02'
collection = '2.2i/runs/DP0.2'
butler = Butler(config, collections=collection)
dataId1 = {'tract': 3441, 'patch': 7, 'band': 'g'}
dataId2 = {'tract': 3442, 'patch': 13, 'band': 'g'}
nImage1 = butler.get('deepCoadd', dataId=dataId1)
nImage2 = butler.get('deepCoadd', dataId=dataId2)
nImage1_wcs = butler.get('deepCoadd.wcs', dataId=dataId1)
nImage1_bbox = nImage1.getBBox()
nImage2_bbox = nImage2.getBBox()
print(nImage1_bbox)
print(nImage2_bbox)
img = Image(Box2I(minimum=Point2I(x=-3900, y=3900), maximum=Point2I(x=4099, y=8099)), dtype=np.float32)
bigImageBBox = img.getBBox()
coaddExposures = [nImage1, nImage2]
dataIds = [dataId1, dataId2]
x = GetTemplateTask()
newimage = x.run(coaddExposures, bigImageBBox, nImage1_wcs, dataIds)
fig = plt.figure(figsize=(24, 12))
display = afwDisplay.Display(frame=fig)
display.scale('asinh', 'zscale')
display.mtv(newimage.template.image)
plt.show()
Results of afwDisplay statement above:
A Zoom in on the seam in DS9: