Over the weekend I investigated the problem of the positional dependence of the PSF model. Unfortunately, I’m still unable to get modelfit_CModel_flux over an entire image, but I’ll summarise what I’ve found:
With characterizeImage.py (and using the config in the previous post) I am able to get modelfit_Cmodel_initial_flux for sources evenly distributed over the entire image, which is great. A small fraction (<5%) return nans, but these seem to be saturated sources, so I think that’s fine.
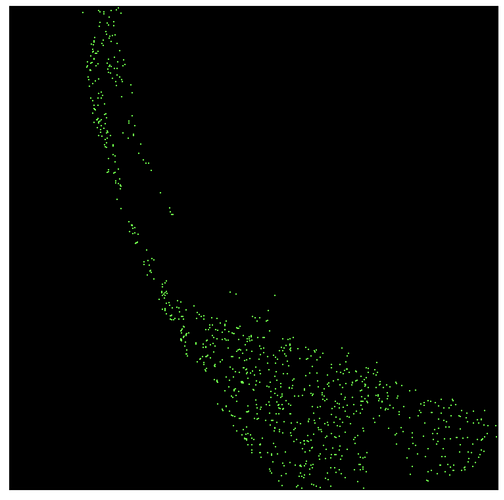
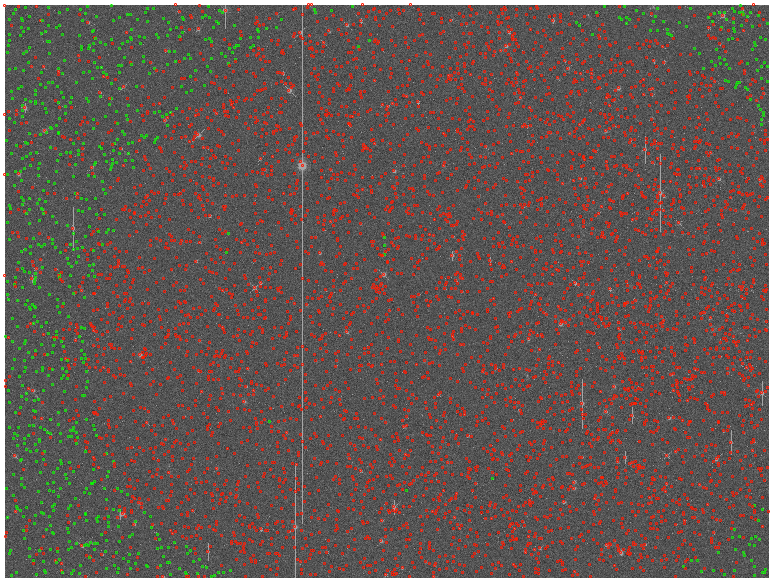
However, modelfit_Cmodel_flux returns large numbers of nans with a strong positional dependence (see below; red points are nans, green are finite fluxes):
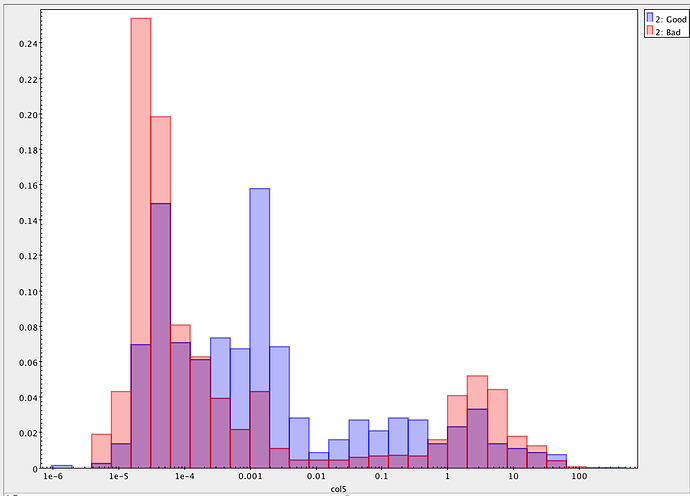
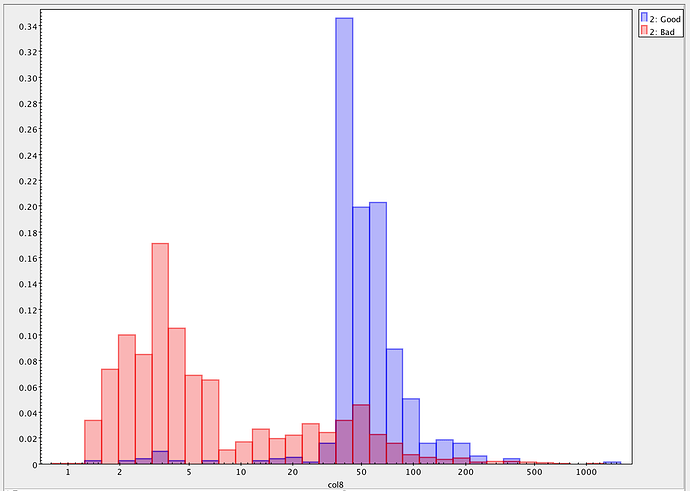
On investigation, all the sources that have nan fluxes have modelfit_CModel_flag_region_maxBadPixelFraction flagged as True, whereas this is False for all sources with finite fluxes. It seems, therefore, that they are failing due some kind of bad pixel mask. However, nothing changes when I either set:
config.measurement.plugins['modelfit_CModel'].region.badMaskPlanes=[]
config.measurement.undeblended['modelfit_CModel'].region.badMaskPlanes=[]
or set
config.measurement.undeblended['modelfit_CModel'].region.maxBadPixelFraction
config.measurement.plugins['modelfit_CModel'].region.maxBadPixelFraction
to either 1, 2, 1000, or None.
I delved into meas_modelfit/include/lsst/meas/modelfit/CModel.h (There Be Dragons!) and came across this comment:
Following the initial fit, we also revisit the question of which pixels should be included in the fit (see CModelRegionControl)
which got me wondering whether modelfit_cmodel_initial is, for some reason, flagging whole regions of my images as bad. However, there doesn’t seem to be a (obvious) flag in the catalog schema that would indicate why this is happening.
Finally, adjusting the config.charImage.measurePsf.psfDeterminer["psfex"] parameters can help a little, but I always have large swathes of sources in my images that return nans for modelfit_cmodel_flux (and the relationship between parameter values and results seems rather chaotic).
So, my questions are:
- Does my suspicion that
modelfit_cmodel_initial is flagging many (contiguous) sources as bad prior to running model_cmodel_dev make sense?
- Does anyone have any thoughts on why this would be the case?
- Similarly, are there any flags that would tell me why
modelfit_cmodel_initial is flagging them as bad?
- Since we suspect this is related to the PSF model, is there some way I can extract or control how the PSF model is interpolated across the image?
- Does anyone have any inspired suggestions as to how this could all be solved?
(I realise that last one is a wildly ambitious question!)
Thanks for helping me get this far, and thanks in advance for any suggestions.