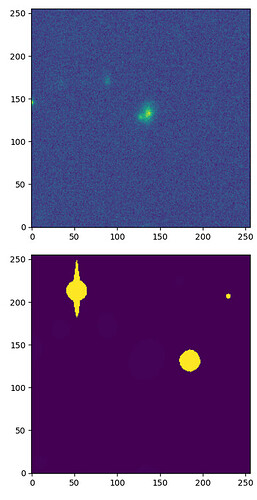
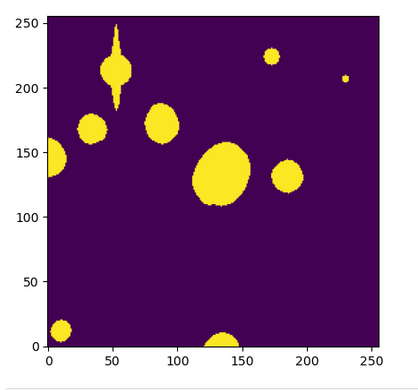
I am working with some visit_image’s and getting cutouts around my object of interest, I noticed in one of the cutouts that the associated mask was wrong (clearly from a totally different part of the image) meanwhile other cutout masks lined up perfectly. I need the masks to flag bad pixels etc, so I might have to just take the full image then manually crop on the full visit_image.mask.arraywhich does seem to be lined up correctly with the visit_image.image.array. I’ve attached a script to reproduce the issue, sorry its not really a “minimal example” but I am still very new so I didn’t want to mess up and lose the one I found that’s misaligned.
from lsst.rsp import get_tap_service
service = get_tap_service("tap")
from lsst.daf.butler import Butler
butler = Butler("dp1", collections="LSSTComCam/DP1")
assert butler is not None
import lsst.geom as geom
import matplotlib.pyplot as plt
import numpy as np
from astropy import units
ra_sn, dec_sn = 58.335054, -48.750303
query = (
"SELECT ra, dec, diaObjectId "
"FROM dp1.DiaObject "
f"WHERE CONTAINS (POINT('ICRS', ra, dec), CIRCLE('ICRS',{ra_sn}, {dec_sn}, {5/60/60})) = 1 "
)
job = service.submit_job(query)
job.run()
job.wait(phases=["COMPLETED", "ERROR"])
print("Job phase is", job.phase)
job.raise_if_error()
assert job.phase == "COMPLETED"
df_sn = job.fetch_result().to_table()
print(f"\nFound {len(df_sn)} source(s)")
diaobjectid = df_sn["diaObjectId"][0]
print(df_sn)
query = (
"SELECT fsodo.coord_ra, fsodo.coord_dec, fsodo.diaObjectId, fsodo.visit, fsodo.detector, fsodo.band, vis.skyRotation, "
"fsodo.tract, fsodo.patch, fsodo.psfDiffFlux, fsodo.psfDiffFluxErr, fsodo.psfFlux, fsodo.psfFluxErr, vis.expMidptMJD "
"FROM dp1.ForcedSourceOnDiaObject as fsodo "
"JOIN dp1.Visit as vis ON vis.visit = fsodo.visit "
f"WHERE fsodo.diaObjectId = {diaobjectid}"
)
job = service.submit_job(query)
job.run()
job.wait(phases=["COMPLETED", "ERROR"])
print("Job phase is", job.phase)
job.raise_if_error()
assert job.phase == "COMPLETED"
df_exposure = job.fetch_result().to_table()
print(f"\nFound {len(df_exposure)} sources")
print(df_exposure)
i = -1 # set to 5 and the mask is aligned correctly!
band = df_exposure["band"][i]
visit = df_exposure["visit"][i]
detector = df_exposure["detector"][i]
tract = df_exposure["tract"][i]
patch = df_exposure["patch"][i]
mjd = df_exposure["expMidptMJD"][i]
dataId = {"band": band, "visit": visit, "detector": detector, "tract": tract, "patch": patch}
# Get calexp, template and sources from butler
visit_image = butler.get("visit_image", dataId=dataId)
# Full image, the mask appears visually aligned
plt.imshow(np.log(visit_image.image.array), origin="lower")
plt.show()
plt.imshow(visit_image.mask.array, origin="lower")
plt.show()
# Cutout, the mask is misaligned
spherePoint = geom.SpherePoint(ra_sn*geom.degrees, dec_sn*geom.degrees)
Radius = 0.01 * units.deg
cutout = visit_image.getCutout(spherePoint, geom.Extent2I(256))
plt.imshow(cutout.image.array, origin="lower")
plt.show()
plt.imshow(cutout.mask.array, origin="lower")
plt.show()